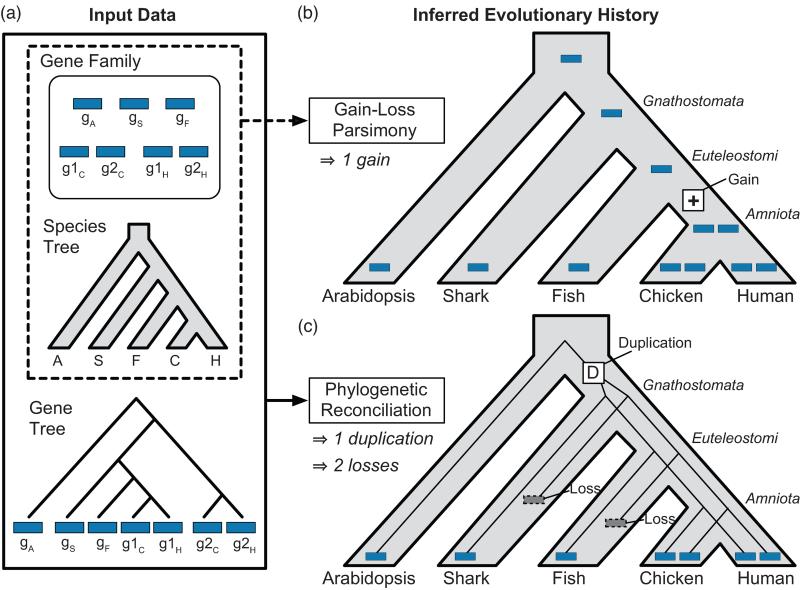
Figure 1. A typical error made by gain-loss methods is avoided with reconciliation.
A gene family with a history of parallel losses illustrates the increased accuracy associated with explicit use of a gene tree by phylogenetic reconciliation. (a) A hypothetical gene family, based on the real enzyme family in Figure 2, possesses one gene in arabidopsis, shark, amphibians, and fish, and two genes in each amniote species. (b) A gain-loss method, Wagner parsimony, incorrectly infers a single gene family member in the common ancestral species and a recent gain on the lineage leading to chicken and human. This scenario implies that all chicken and human genes are equally related to gS and gF, an inference that is not supported by the true tree. (c) Gene tree-species tree reconciliation correctly infers an earlier duplication, followed by parallel losses in the shark and fish lineages, and shows that g1H and g1C are more closely related to gS in shark and gF in fish, than to g2H and g2C.