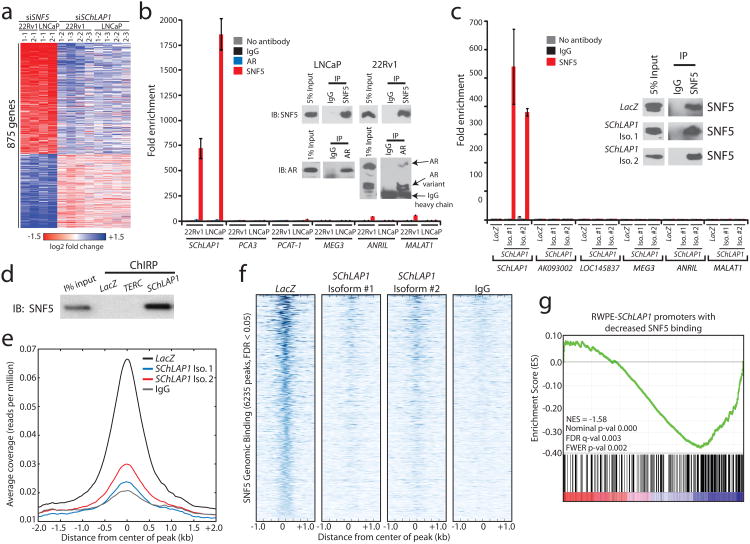
Figure 4. SChLAP1 antagonizes SNF5 function and attenuates SNF5 genome-wide localization.
(a) Heatmap results for SChLAP1 or SNF5 knockdown in LNCaP and 22Rv1 cells. (b) RNA immunoprecipitation (RIP) of SNF5 or AR in SNF5 in 22Rv1 and LNCaP cells. Inset Western blots demonstrate pulldown efficiency. (c) RIP analysis of SNF5 in RWPE cells overexpressing LacZ, SChLAP1 isoform #1, or SChLAP1 isoform #2. Inset Western blots demonstrate pulldown efficiency. (d) Pulldown of SChLAP1 RNA using Chromatin Isolation by RNA Purification (ChIRP) recovers SNF5 protein in RWPE-SChLAP1 isoform 1 cells. LacZ and TERC serve as controls. (e) A global representation of SNF5 genomic binding over ±2kb window surrounding each SNF5 ChIP-Seq peak in RWPE-LacZ, RWPE-SChLAP1 isoform 1, and RWPE-SChLAP1 isoform 2 cells. (f) A heatmap representation of SNF genomic binding at target sites in RWPE-LacZ, RWPE-SChLAP1 isoform 1, and RWPE-SChLAP1 isoform 2 cells. A ±1kb interval surrounding the called SNF5 peak is shown. (g) Gene set enrichment analysis results showing significant enrichment of ChIP-Seq promoter peaks with >2-fold loss of SNF5 binding for underexpressed genes in RWPE-SChLAP1 cells.