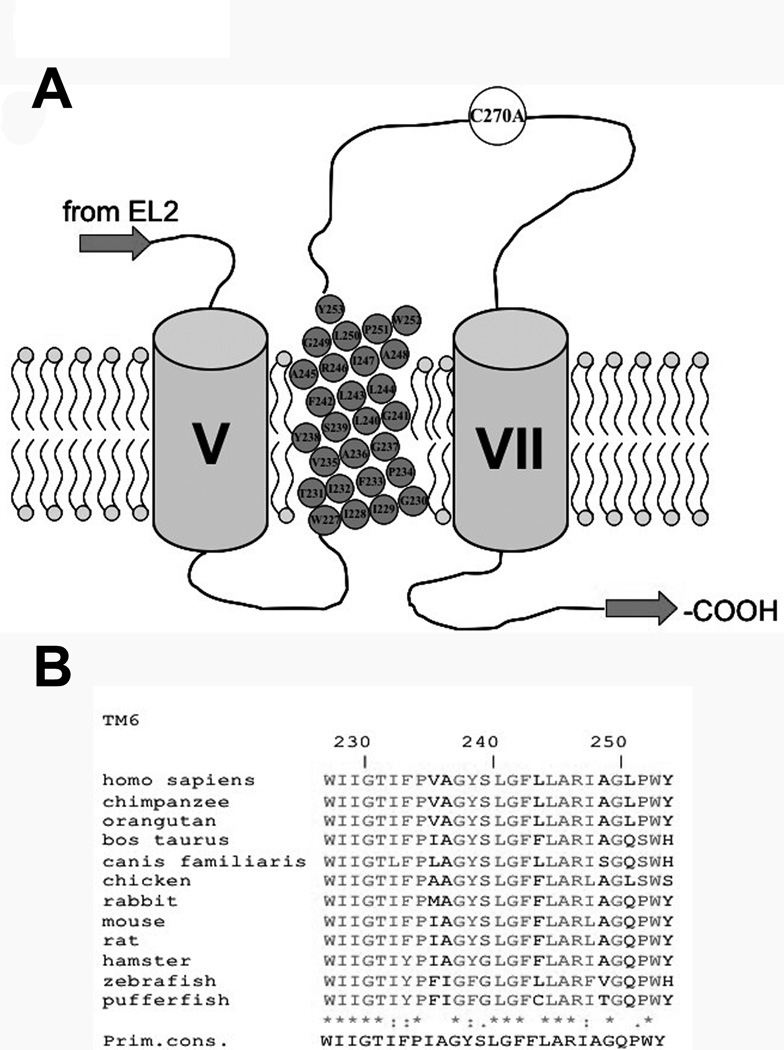
Figure 1. Multiple sequence alignment of TM6 amino acids.
[A] Secondary structure model of the last three transmembrane domains (TMD) of hASBT according to the 7TM model. Roman numerals indicate flanking TMD while TMD 6 amino acids are represented by gray circles inscribed with amino acid identity and position. Phospholipids of plasma membrane represented by circle (polar phosphate head group) with two tails (hydrophobic lipids). Top is exofacial, bottom cytosolic. [B] Sequence alignment of amino acids 227–253, putatively forming TM6, for all known ASBT paralogs. Sequences were retrieved from GeneBank and aligned via the MULTALIN routine with annotation performed via MPSA. Shaded regions denote complete amino acid conservation among all species. Amino acid positioning relative to human ASBT is indicated by numbering on top. Bottom line indicates primary consensus for the TM6 region.