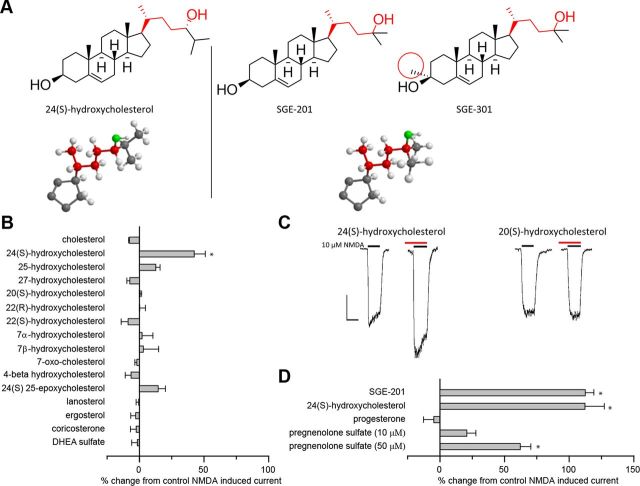
Figure 1.
24(S)-HC and SGE-201 are potent oxysterol positive allosteric modulators of NMDA receptors. A, Natta projection structures for 24(S)-HC, SGE-201, and SGE-301. Note that the key hydroxyl group (denoted in red) in all three structures is on the same carbon relative to the cholesterol backbone. The similarity in the location of the hydroxyl group is further emphasized in the three dimensional stick and ball models below (D-ring and C-17 side chain shown). The 3-α methyl group that distinguishes SGE-201 and SGE-301 is circled in red. B, Effects of endogenous oxysterols and other cholesterol metabolites on NMDA receptor currents. Cultured primary hippocampal neurons were preincubated with 10 μm test compound in 0.5 μm glycine for 90 s, followed by 10 s NMDA (10 μm). The percentage change in NMDA current is plotted. C, Representative traces from B. The red lines represent application of test article (note that the red lines do not encompass the full 90 s of preincubation). The black lines represent the application of NMDA (10 μm, 10 s). Scale bar: vertical = 200 pA, horizontal = 10 s. D, Active oxysterols compared with pregnenolone metabolites. All compounds were tested at 10 μm, 90 s preincubation, with the exception of 24(S)-HC (10 μm, 360 s preincubation), and pregnenolone sulfate (10 and 50 μm, 90 s preincubation). Cholesterol was solubilized in ethanol rather than DMSO. Asterisk represents a significant difference from current induced by NMDA alone (p < 0.05).