Abstract
Bone is a dynamic tissue that is constantly renewed by the coordinated action of two cell types, i.e., the bone-resorbing osteoclasts and the bone-forming osteoblasts. However, in some circumstances, bone regeneration exceeds bone self repair capacities. This is notably often the case after bone fractures, osteolytic bone tumor surgery, or osteonecrosis. In this regard, bone tissue engineering with autologous or allogenic mesenchymal stem cells (MSCs) is been widely developed. MSCs can be isolated from bone marrow or other tissues such as adipose tissue or umbilical cord, and can be implanted in bone defects with or without prior amplification and stimulation. However, the outcome of most pre-clinical studies remains relatively disappointing. A better understanding of the successive steps and molecular mechanisms involved in MSC-osteoblastic differentiation appears to be crucial to optimize MSC-bone therapy. In this review, we first present the important growth factors that stimulate osteoblastogenesis. Then we review the main transcription factors that modulate osteoblast differentiation, and the microRNAs (miRs) that inhibit their expression. Finally, we also discuss articles dealing with the use of these factors and miRs in the development of new bone MSC therapy strategies. We particularly focus on the studies using human MSCs, since significant differences exist between osteoblast differentiation mechanisms in humans and mice for instance.
Keywords: Mesenchymal stem cells, Osteogenesis, Runt-related 2, Wnt, MicroRNAs
Core tip: Several excellent reviews on the transcription factors involved in osteoblast differentiation have recently been published, but none also presented the microRNAs (miRs) that control the expression of these transcription factors. Moreover, most of these reviews mainly reported mouse studies but important differences are well acknowledged between humans and mice. For instance vitamin D3, an important hormone controlling bone homeostasis, has very different effect in these species. Therefore, in the present review we particularly focus on human cells to present the transcription factors and miRs controlling mesenchymal stem cells-osteoblastic differentiation.
BONE REPAIR WITH MESENCHYMAL STEM CELLS
Historically, Friedenstein et al[1] were the first to report the presence of fibroblastoid cells in the adult bone marrow that can make bone and reconstitute a hematopoietic microenvironment when transplanted subcutaneously. These mesenchymal stem cells (MSCs) were later reported to contribute to various musculoskeletal tissues such as bone, cartilage, fat, muscle, ligament and tendon[2]. In 2006, the International Society for Cellular Therapy proposed that cells with the following characteristics should be considered as MSCs, (1) cells adherent to plastic in culture; (2) presence of CD105, CD73 and CD90 but absence of CD34, CD45, CD14 or CD11b, CD79α or CD19, and HLA-DR molecules; and (3) cells with the capacity to differentiate into osteoblasts, chondrocytes and adipocytes[3]. However, although these criteria are widely accepted, they may still be imperfect. Indeed, the three markers are co-expressed in a wide variety of cells, and may therefore not be able to indentify a single MSC population in vivo[4].
MSCs represent less than 0.01% of the bone marrow cell population. At birth, the frequency of MSCs has been reported as 1 MSC/104 BM-mononuclear cells, decreasing to 1 MSC/105 BM-mononuclear cells in teenagers to 1 MSC/2 × 106 BM-mononuclear cells in 80-year-old individuals[5]. To overcome the drawbacks associated with MSC isolation from bone marrow, other sources have been contemplated. MSCs can indeed be recovered from several different locations such as adipose tissue[6], dental pulp[7] and umbilical cord[8]. Recently, Sacchetti et al[9] reported CD146 high pericytes surrounding bone marrow vascular sinusoids can be considered as MSCs as they are self-renewing osteoprogenitors capable of ectopic bone formation. Finally, differences appear to exist between MSC populations from different tissues, which represents an additional challenge to devise a universal definition[10].
MSC differentiation into osteoblasts can be achieved by adding vitamin D3, ascorbic acid and β-glycerophosphate to the culture medium[11]. Several laboratories use dexamethasone, a synthetic glucocorticoid, instead of vitamin D3. Dexamethasone appears to optimize differentiation from MSCs, but not specifically to the osteoblast lineage[12]. In osteogenic conditions, human MSCs secrete a matrix enriched in type I collagen which will be be mineralized with apatite crystals upon activation of tissue-non specific alkaline phosphatase (TNAP) (Figure 1)[13]. Osteoblasts also secrete a tissue-specific protein, osteocalcin, recently shown to act as a circulating hormone involved in the control of insulin secretion and sensitivity[14]. However, although this protein is a useful marker of osteoblast differentiation, it doesn’t seem to impact bone formation. Eventually, some osteoblasts will become surrounded by a mineralized collagen matrix and further differentiate into bone-residing osteocytes, which secrete different proteins such as sclerostin, a canonical Wnt signaling inhibitor, and dentin matrix protein-1, a molecule controlling phosphatemia[15].
Figure 1.
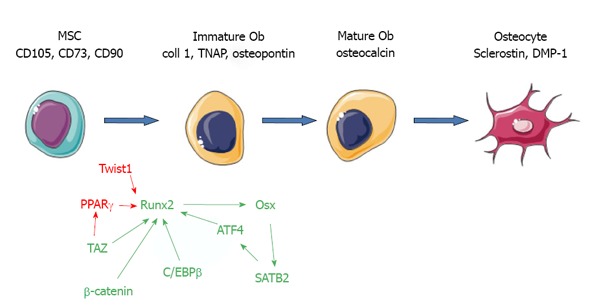
transcription factors involved in osteoblast differentiation form mesenchymal stem cells. Markers of differentiation are shown in black, stimulatory transcription factors in green, and inhibitory ones in red. MSCs: Mesenchymal stem cells; ATF4: Activating transcription factor 4; C/EBP: CCAAT/enhancer-binding proteins; Coll: Collagen; DMP-1: Dentin matrix protein-1; Ob: Osteoblast; Osx: Osterix; PPAR: Peroxysome proliferator activated receptor; SATB2: Special AT-rich sequence binding protein 2; TAZ: Transcription coactivator with binding capacity to PDZ motifs; TNAP: Tissue-nonspecific alkaline phosphatase; Runx2: Runt-related 2.
MSCs have been implanted in association with different scaffolds to rebuild bone[16,17]. Injection of MSCs has also been shown to correct bone defects. Notably, allogenic bone marrow transplants or injection of isolated MSCs in children with osteogenesis imperfecta (OI) have improved bone formation and function[18,19]. However, although promising data were reported, many others led to contrasting if not disappointing results[20]. In this regard, it appears crucial to better understand the molecular mechanisms of osteoblast differentiation from human MSCs. This will allow us to improve the bioactivity of injected MSCs or MSC-containing hybrid materials by stimulating their osteoblast differentiation. This may be achieved through genetic modification of MSCs. For instance, autologous MSCs may be modified to correct the abnormal collagen synthesis in patients with OI[21]. Several excellent reviews on osteoblast differentiation have been published in recent years. To our knowledge however, none has focused on the interactions between transcription factors and microRNAs in human mesenchymal stem cells specifically. We believe that it is particularly important since significant differences are well acknowledged between osteoblastogenesis of human and mouse MSCs. For instance, while vitamin D3 binds to a vitamin D response element (VDRE) in the osteocalcin promoter in humans and rats, the mouse osteocalcin promoter is devoid of any VDRE and vitamin D3 exerts an indirect inhibitory effect on osteocalcin transcription[22,23].
GROWTH FACTORS STIMULATING MSC-OSTEOBLASTIC DIFFERENTIATION
Two families of growth factors appear to stimulate osteoblast differentiation from MSCs: the Wnt (a portmanteau of Wingless and integration 1) family and the bone morphogenetic proteins (BMPs).
Wnt family members
Wnt proteins are a family of 19 highly conserved secreted glycoproteins that play essential roles during development and tissue homeostasis[24]. Some Wnt proteins such as Wnt3a and Wnt10b bind to Frizzled receptors, and recruit the LRP5/6 coreceptors to activate the canonical signaling pathway, leading to glycogen synthase kinase-β inhibition, β-catenin stabilization, translocation into the nucleus and regulation of T-cell factor/lymphoid enhancer factor (TCF/LEF) transcriptional activity. Binding of Wnt proteins to LRP5/6 is inhibited by secreted factors such as Dickkopf-related protein 1 (Dkk1)[24]. Dkk1 binds to LRP5/6 causing the receptor to attract Kremen, and this interaction promotes clathrin-mediated internalization thereby inactivating LRP5/6.
The importance of the canonical Wnt signaling in bone is well-acknowledged. Genetic reports established that Wnt/β-catenin activity is essential for bone development[25]. Deficiency of Dkk1 is associated with increased bone formation in mice and humans[26]. Wnt10b may be particularly important for bone formation. Wnt10b is expressed in the bone marrow by osteoblast progenitors[27], and transgenic overexpression of Wnt10b in mesenchymal cells leads to increased bone density and accelerated osteoblastogenesis in vitro, whereas Wnt10b-/- mice have reduced trabecular bone[28]. Moreover, Wnt10b seems to stimulate osteoblast functions through a positive autocrine loop[29]. On the other hand, other recent findings indicate that canonical Wnt signalling inhibits osteoblast differentiation in human MSC cultures[30-32]. These contrasting findings have been reconciled recently by Liu et al[33] who found that Wnt/β-catenin signalling favours osteogenic commitment in basal medium by inhibiting MSC commitment into adipocytes, but inhibits osteoblast differentiation in osteogenic conditions. This was confirmed by Kang et al[34] who reported that Wnt10b induction of osteogenesis in mouse progenitors was due to inhibition of peroxysome proliferator-activated receptor (PPAR)γ and CCAAT/enhancer-binding protein (C/EBP)α activity. The mutual inhibition between β-catenin and PPARγ will be discussed below.
Alternatively, non-canonical Wnt members may also be involved in the effects of TNF-α on ossification. In particular, Wnt5a seems to be the predominant Wnt variant expressed during osteoblastic differentiation of human MSCs[35]. Wnt5a+/- mice present a reduced bone mass phenotype with decreased osteoblast number[36]. Wnt5a appears to stimulate osteoblast differentiation through an autocrine loop in human MSCs[37,38]. Another non-canonical Wnt with a potential interest in bone repair is Wnt4. In two different models of craniofacial bone injury, Chang et al[39] observed that human MSCs genetically engineered to express Wnt-4 enhanced osteogenesis and improved the repair of craniofacial defects in nude mice.
Bone morphogenetic proteins
BMPs are growth factors that belong to the transforming growth factor beta (TGF-β) superfamily[40,41]. The term, bone morphogenetic protein was first introduced to describe the components in demineralized bone matrix that can induce ectopic bone formation when implanted intramuscularly or subcutaneously into rodents[42,43]. To date, more than 20 BMP members have been characterized. As TGF-β, BMPs trigger cellular responses mainly through the Smad pathway[44], although they can also activate the mitogen-activated protein kinase pathway[45]. In the Smad pathway, type II and type I receptors with serine/threonine kinase activity and intracellular Smad proteins relay the signal from the cell surface to the nucleus. Three type II receptors can bind BMPs: type II BMP receptor, and type II and IIB activin receptors (ActR-II and ActR-IIB)[40]. Three type I receptors for BMPs have also been characterized: type IA and IB receptors (BMPIA or ALK3 and BMPIB or ALK6), and type IA activin receptor (ActRIA or ALK2). The receptors activated by ligand binding phosphorylate a subgroup of receptor-regulated Smads (R-Smads including Smad 1, 5 and 8). The phosphorylated R-Smads then disassociate from their receptor and form complexes with the common partner Smad 4. Smad heterodimers then migrate into the nucleus where they associate with transcription factors to regulate gene transcription. This Smad signal is inhibited by Smad 6 and Smad 7, which block phosphorylation of R-Smads.
BMP factors are important in skeletogenesis[40]. BMP-2 is expressed in areas surrounding cartilage condensations[46,47], while BMP-4 is expressed in perichondrium[47]. BMP-2 is also expressed in periosteal and osteogenic zones[46]. Due to their effect on runt-related 2 (Runx2) and osterix expression[48], BMPs are very potent inducers of mesenchymal progenitor cell differentiation into osteoblasts[49]. Recombinant BMPs can be added in different materials such as in collagen sponges and calcium phosphate ceramics to be delivered in situ for clinical practice[50-52]. In humans, recombinant human BMP-2 and BMP-7 have been approved for clinical use in orthopedic surgery for long bone open-fractures treated with intramedullary fixation and non-union fractures, and in spine surgery for spinal fusion in place of iliac crest bone graft[53]. BMPs do not seem to accelerate fracture healing but tend to increase healing rates without requiring a secondary procedure[54]. Nevertheless, several concerns today complicate the use of BMPs, such as heterotopic ossifications, immunogenic reactions or hardware failure[54,55]. Moreover, the clinical interest of BMPs is limited to local applications, and BMPs may not represent an alternative treatment to systemic bone diseases such as osteoporosis. Systemic use of BMPs is limited by their non-skeletal effects, mitogenicity, and short half-life.
TRANSCRIPTION FACTORS INVOLVED IN MSC-OSTEOBLAST DIFFERENTIATION
Stimulatory transcription factors
β-catenin: As detailed above, β-catenin is potently activated in the canonical Wnt signaling pathway[24]. In this pathway, unphosphorylated β-catenin molecules accumulate in the cytoplasm, translocate to the nucleus, and activate the transcription of downstream genes by binding tLEF/TCF transcription factors. Conditional deletion of β-catenin gene in Dermo-Cre or Prx1-Cre transgenic mice reveals its essential role in osteoblast differentiation[25,56]. In addition, conditional deletion of β-catenin gene in Wnt1-Cre transgenic mice, in which Cre is expressed in neural crest cell precursors, results in loss of cranial bones derived from neural crest cells[57]. Interestingly Runx2 is expressed in β-catenin deficient cells[25,56], but is strongly enhanced by β-catenin/TCF1. It is required for osterix expression and osteoblast differentiation[58] (Figure 1).
Runx2: Runx2 belongs to the Runx family, which consist of Runx1, Runx2 and Runx3. These transcription factors form heterodimers with Cbfb and bind to the consensus sequence TGPyGGPyPy[59]. Runx2 is considered as the master osteoblast transcription factor (Figure 1). It was identified as a factor binding to an osteoblast specific cis-acting element in the promoter of the genes encoding for osteocalcin[60]. Runx2 deficiency in mouse leads to the formation of a skeleton devoid of osteoblasts[61,62]. In man, inactivating mutations in Runx2 leads to a skeletal dysplasia called cleidocranial dysplasia[63]. Runx2 regulates many genes that determine the osteoblast phenotype. Runx2 is sufficient to induce the expression of many osteoblast markers, such as osteocalcin, in non-osteoblastic cells[60]. However, Runx2 overexpression in osteoblasts severely reduces osteocalcin expression and osteoblast maturation[64,65]. Therefore, whereas Runx2 is required to commit undifferentiated cells towards the osteoblast lineage, it appears to maintain these cells in an immature stage[66].
In murine fibroblasts, the forced expression of Runx2 is sufficient to induce expression of osteoblast markers such as collagen type I, osteocalcin or bone sialoprotein. Adenoviral overexpression of Runx2 in mouse MSCs generated substantially more bone than control MSCs when implanted in subcutaneous tissue or in calvarial defects[67]. Similarly, rat bone marrow stromal cells transduced with Runx2 retroviral vector seeded onto 3D-fused deposition-modeled polycaprolactone scaffolds, produced biologically-equivalent mineralized matrices at nearly 2-fold higher rates than control cells[68]. In human MSCs isolated from adipose tissue, electroporation of Runx2 stimulated osteoblast differentiation in vitro with increased expression of alkaline phosphatase and osteocalcin[69].
Transcription coactivator with binding capacity to PDZ motifs: Transcription coactivator with binding capacity to PDZ motifs (TAZ) was originally identified during a series of control experiments in a proteomic screen looking for 14-3-3-interacting proteins[70]. TAZ contains a 14-3-3-binding motif, a single WW domain, an extended coiled-coiled region within a larger transcriptional regulatory domain, multiple sites of phosphorylation, and a C-terminal motif that can interact with PDZ-containing proteins[71]. The WW domain of TAZ binds to the sequence motif Pro-Pro-X-Tyr. This motif can be found within the regulatory regions of a large number of transcription factors, including Runx2 and PPARγ, as well as members of the Sox, and SMAD families, suggesting that TAZ may be involved in the regulation of MSC commitment and differentiation into osteoblasts, adipocytes and chondrocytes[71]. The WW domain-containing molecule TAZ directly interacts with Runx2 and co-activates Runx2-dependent gene transcription[72]. In contrast, TAZ binds to, and markedly inhibits, the ability of PPARγ to drive the expression of adipocyte-associated genes such as adipocyte protein 2, and depletion of TAZ increases their adipocyte differentiation[72]. The processes through which TAZ is induced and/or activated are poorly understood[71]. TAZ levels increase substantially in MSCs induced to differentiate into osteoblasts with BMP-2, whilst conversely, they decrease during adipocyte differentiation[71]. It was also reported that TNF-α stimulates osteogenesis in hMSCs from adipose tissue through NF-κB activation and TAZ expression[73]. However, the pathophysiological significance of this finding remains obscure. In mouse mesenchymal cells, high-throughput screening allowed to identify a chemical compound, so-called TM-25659, that enhances TAZ nuclear localization and osteoblast differentiation at the expense of adipocytes[74]. Moreover, TM-25659 suppressed bone loss in vivo and decreased weight gain in an obesity model. Although this compound seems to have a favorable pharmacokinetic profile, work remains to be done to demonstrate its possible interest in clinical application.
Special AT-rich sequence binding protein 2: Special AT-rich sequence binding protein 2 (SATB 2) is a member of the family of special AT-rich binding proteins that binds to nuclear matrix attachment regions (MARs) and activates transcription in a MAR-dependent manner. SATB2 inactivation in man results in cleft palate[75]. SATB 2-/- osteoblasts are characterized by a decreased differentiation, illustrated by reduced bone sialoprotein (BSP) and osteocalcin expression[76]. SATB2 can physically interact with both activating transcription factor 4 (ATF4) and Runx2 and enhance the transactivation function of both proteins[76]. Overexpression of SATB 2 in mouse bone marrow stromal cells stimulates expression of osterix and BSP[77]. Transplanted SATB 2-overexpressing adult stem cells genetically double-labeled with BSP promoter-driven luciferase and β-actin promoter-driven enhanced green fluorescent protein into mandibular bone defects accelerated new bone formation[77]. In addition, SATB 2-overexpressing murine induced pluripotent stem cells[78] show increased mineral nodule formation and elevated mRNA levels of key osteogenic genes, osterix, Runx2, Bsp and osteocalcin[79]. SATB 2-overexpressing induced pluripotent stem cells combined with silk scaffolds and transplanted into critical-size calvarial bone defects created in nude mice induced enhanced bone repair[79].
Osterix: Besides Runx2, the second transcription factor absolutely required for osteoblast differentiation is Osterix (Osx, also known as Sp7). Osx is a zinc finger-containing transcription factor belonging to the SP family of transcription factors. Osx is specifically expressed in osteoblasts, and is required for bone formation[80]. The fact that Runx2 is expressed in Osx-deficient mice combined with the absence of Osx in Runx2 null mice places Osx downstream of Runx2[80]. Actually, Runx2 may induce Osx expression, through direct binding on its promoter[81]. Interestingly, Osx binds to the promoter of Satb 2 to increase the transcription of the Satb 2 gene[82]. Thus, part of the effects of Osx may rely on SATB 2 activity. Murine bone marrow stromal cells overexpressing Osx associated with type I collagen sponge as a carrier exhibited five times more amounts of newly formed calvarial bone than that the control group in adult mice[83]. In addition, overexpression of Osx in human umbilical cord-derived MSCs result in increased alkaline phosphatase activity and osteocalcin expression, and enhanced bone regeneration in nude mice using polylactic-co-glycolic acid as a carrier[84].
Smads: Runx2 cooperates with Smad (a portmanteau of Sma in Drosophila and Mad in C. elegans) 2 and Smad 5 to regulate bone-specific genes[85,86]. These interactions appear to be important in vivo[87-89]. Whilst Runx2 alone does not induce osteoblast differentiation, it synergizes with Smad 2 and Smad 5 to achieve this event. Mutant Runx2 with a truncated transcription activation domain fails to interact with Smad1 and consistently blocks BMP/Smad-induced osteoblast differentiation[86]. In addition to Runx2, menin, the product of the multiple endocrine neoplasia type 1 gene, is required for BMP-induced osteoblast differentiation[90]. Menin interacts with both Runx2 and Smad 1/5 in multipotential mesenchymal cells. When menin is knocked down, the cells fail to differentiate into the osteoblast lineage.
CCAAT/enhancer-binding proteins β: CCAAT/enhancer-binding proteins (C/EBPs) belong to the group of basic leucine zipper transcription factors. They are known to modulate both adipocyte and osteoblast differentiation. C/EBPβ forms a homodimer or heterodimer complex with other C/EBP family members. C/EBPβ is expressed before PPARγ and induces it[91,92]. More precisely, two main protein forms of C/EBPβ, induced by alternative translation initiation, present opposite effects on adipogenesis[91]. Whereas LAP, the main long isoform, is proadipogenic, the short one, LIP, acts as a dominant negative inhibitor of LAP. In murine mesenchymal cells, LIP inhibits adipocyte differentiation and preferentially induces osteoblast differentiation[93]. C/EBPβ promotes osteoblast differentiation of mesenchymal cells in Runx2-dependent and -independent mechanisms[93]. C/EBPβ up-regulates Runx2 expression by directly binding to the Runx2 P1 promoter in mesenchymal, pre-osteoblastic, and osteoblastic cells[94]. In addition, C/EBPβ interacts with Runx2 and activates the transcription of the osteocalcin gene[95]. C/EBPβ heterodimerizes with activating transcription factor 4 (ATF4, presented below), another basic leucine zipper transcription factor crucial for osteoblast maturation. This complex transactivates osteocalcin-specific element 1 of the osteocalcin promoter[96]. Absence of all C/EBPβ isoforms results in decreased bone mass in mice, associated with impaired osteoblast differentiation and functional deficiency[96]. These data suggest that C/EBPβ activates osteoblastogenesis. However, before commitment C/EBPβ may act as a transcriptional repressor of Runx2 and of osteoblast differentiation[91,97]. Mechanistically, it has been proposed that once osteogenic differentiation is initiated, Smad3 expression increases, binds to C/EBPβ, and blocks its inhibitory action on Runx2[98].
Activator protein 1 proteins: Activator protein 1 represents heterodimeric transcription factors composed of members of the Jun and Fos family of basic leucine zipper proteins. Overexpression of ΔFosB or Fra1 leads to enhanced bone formation. Osteopetrosis in ΔFosB overexpressing mice is due to the inhibition of mesenchymal cell differentiation into adipocytes, leading to an increased number of osteoblasts[99]. Moreover, conditional Fra1-/- mice display reduced levels of several matrix proteins, such as osteocalcin[100]. Finally conditional deletion of JunB causes bone defects with reduced osteoblast proliferation, and expression of osteocalcin and bone sialoprotein[101].
ATF4: Mice deficient in ATF4 display a decreased bone formation, leading to a severe low bone mass phenotype[102]. At the molecular level, ATF4 directly binds to the promoter of osteocalcin to activate transcription[102]. This activation appears to rely on the physical interaction between ATF4, SATB2 and Runx2 at the promoter level[103]. ATF4 may also cooperate with C/EBPβ to activate transcription of the osteocalcin gene[96]. Finally, ATF4 also plays indirect effects through its activation of amino acid transport[104]. Indeed, osteoblasts from ATF4-/- mice do not synthesize normal levels of typeIcollagen unless nonessential amino acids are added to the culture[102].
Inhibitory TFs
Peroxysome proliferator-activated receptor γ: PPARγ proteins are expressed in mice and humans as two different isoforms, PPARγ1 and PPARγ2, due to alternative promoter usage and alternative splicing. PPARγ1 is ubiquitously expressed whereas PPARγ2 expression is restricted to adipocytes[105,106]. Homozygous PPARγ-deficient ES cells fail to differentiate into adipocytes, but spontaneously differentiate into osteoblasts[107]. Heterozygous Pparγ-deficient mice exhibit a high bone mass phenotype but normal osteoblast functions[107]. Pparγ2 has been reported to bind to Runx2 and inhibit its transcriptional activity[108]. Inhibition of PPARγ with the pharmacological inhibitor GW9662 in human MSCs stimulates mineralization and bone formation in vitro and in vivo[109,110]. Besides the inhibition of Runx2, PPARγ inhibitory effects may also include β-catenin. Indeed, activated PPARγ in mesenchymal cells induces the proteasomal degradation of β-catenin following direct interaction[111]. Moreover, Lu et al[112] reported that the PPARγ inhibitor GW9662 significantly activates TCF reporter plasmid activity. Furthermore, Krause et al[110] reported that GW9662 treatment of hMSCs resulted in β-catenin accumulation in the nucleus and PPARγ nuclear export. However, it was recently suggested that whereas PPARγ2 pro-adipocytic activity relies on β-catenin inhibition, its anti-osteoblastic activity is independent of this interaction[113].
On the other hand, a stimulatory role for PPARγ in osteoblast differentiation has been reported. Overexpression of PPARγ2 in C3H10T1/2 mouse mesenchymal precursors do not only promote adipogenic differentiation, but also enhances osteogenic differentiation upon BMP-2 stimulation[114]. Conversely, MSCs with PPARγ2 knockdown or mouse embryonic fibroblasts derived from PPARγ2-/- mice exhibit a decrease in adipocyte differentiation, coupled with reduced osteoblastogenesis and decreased mineralization[114]. In mouse MC3T3-E1 osteoblasts, activation of PPARγ1 with low doses of agonists stimulated alkaline phosphatase activity and mineralization[115]. In hMSCs, two PPARγ antagonists, BADGE and GW9662, as well as lentiviral knockdown of PPARγ inhibited adipogenesis but had no effect on osteoblastogenesis[116].
In conclusion, while most data seem to demonstrate an inhibitory effect of PPARγ on osteoblastogenesis, several articles suggest that PPARγ action on osteoblasts may actually be more ambiguous. Several mechanisms may account for these discrepancies. For instance, PPARγ directly binds and inhibits Runx2[108], and therefore inhibits MSC commitment into osteoblasts. On the other hand, since Runx2 appears to maintain osteoblasts in an immature stage[66], PPARγ may participate in osteoblast maturation. Besides Runx2, PPARγ has also been shown to bind and inhibit β-catenin pro-osteogenic function[111]. However, β-catenin and PPARγ may not be systematically inhibitory because an elegant article recently showed that BMP-2 activated β-catenin/PPARγ dimers have their specific transcriptional targets in endothelial cells[117]. Since BMP-2 is a potent osteogenic factor, PPARγ roles in osteoblasts may therefore be more subtle than commonly accepted.
Finally, PPARγ activity is also dependent of a wide number of factors, such as 1,25(OH)2 vitamin D3 receptor, PPAR coactivator (PGC-1), the histone acetyltransferase p300, CREB binding protein, and steroid receptor coactivator-1[118]; its effects on osteoblasts may thus vary as a function of cell differentiation, species and mode of activation or inactivation. For instance, it was suggested that full but not partial agonist activation inhibits expression of osteoblast markers in human MSCs[119].
Twist1: In mouse, there is a 4-5 d delay between the appearance of Runx2 and that of its target, osteocalcin. This delay seems to be due to the co-expression of Twist1[120]. Twist1 is a basic helix-loop-helix transcription factor. Haploinsufficiency at the Twist1 locus causes Saethre-Chotzen syndrome, a form of craniosynostosis, i.e., an increase in bone formation in the skull[121,122]. Molecularly, Twist1 binds to the DNA binding domain of Runx2, and inhibits its transcriptional activity. Similarly, Twist1 also interacts with ATF4 and decreases its binding to the Osteocalcin promoter[123]. As a consequence, osteoblast differentiation during development proceeds when and where Twist1 expression drops. In C3H10T1/2 mouse cell progenitors, silencing of Twist1 using short hairpin RNA expression enhanced osteoblast gene expression and matrix mineralization in vitro[124]. In human MSCs, overexpression of Twist1 and Dermo-1 was associated with a decrease in the gene expression of osteoblast-associated markers, bone morphogenic protein-2, bone sialoprotein, osteopontin, alkaline phosphatase and osteocalcin[125].
MICRORNAS INVOLVED IN MSC OSTEOBLASTIC DIFFERENTIATION
MicroRNAs
MicroRNAs (miRs) are small (19-23 nt) endogenous non-coding single-stranded RNA transcribed from both intergenic and genic regions of the genome[126,127]. They are highly conserved molecules that control gene expression post-transcriptionally by binding to the 3′UTR of target mRNA. Near-perfect complementarity between the sequence of miR and its target results in the cleavage of target mRNA, whereas partial complementarity results in its translational inhibition[128]. The biogenesis of these small regulatory RNA molecules starts out as primary transcripts termed pri-miR. The pri-miR is first processed in the nucleus by the RNAse III enzyme DROSHA to produce pre-miRNAs. Once in the cytoplasm, pre-miRs are further processed by a second RNase III enzyme, DICER1 resulting in dsRNA miR complex, which unwound by the helicase activities of the Argonaute multiprotein complex known as the RNA-induced silencing complex (RISC). The preferred guide strand is incorporated into the RISC complex[129]. MiR expression has both spatial and temporal specificity as well as tissue or cell specificity[130]. Strikingly, bioinformatics analysis suggests that up to 30% of human genes may be regulated by miR[131]. MiRs act as key regulators in diverse biological processes, such as early development, cell proliferation, differentiation, apoptosis, cancer and have the potential to control the expression of virtually any gene[132]. Some miRs are directly involved in the formation of the human skeletal system. Thus, miRs have the great potential to become a research focus for the prevention and treatment of skeletal diseases[130].
MiRs and osteoblast differentiation
Conditional deletion of the miR processing enzyme Dicer in osteoblasts, chondrocytes, and osteoclasts has revealed their essential role in normal skeletal development and bone homeostasis[133]. Differential expression of miRs has a major impact on the regulation of osteoblast differentiation[134], where by various signaling pathways/transcription factors responsible for osteoblast differentiation can be modulated by miRs. An increasing number of miRs have been identified to negatively regulate osteoblast differentiation and bone formation by targeting important osteogenic factors and positively affect it by targeting negative regulators of osteogenesis.
Negative regulators: Many miRs were shown to act as inhibitors of osteoblast differentiation (Table 1). These include miR-206 by targeting connexin 43 gene (Cx43)[135] or MiR-34 that decreases SATB2 accumulation[136]. Additionally, Hsa-miR-27a and has-miR-489 down-regulate differentiation through repression of TNAP expression[137]; miR-204 a negative regulator of Runx2 inhibits osteogenesis and promotes adipogenesis of mesenchymal progenitor cells and BMSCs[138]. MiR-133 and miR-135 target Runx2 and Smad1/5 respectively in C2C12 mouse mesenchymal progenitors[139]. MiR-433 suppresses BMP2-induced osteoblast differentiation via direct targeting of Runx2 mRNA in C3H10T1/2 cells[140]. Finally, some under-expressed miRs (hsa-miR-31, hsa-miR-106a, hsa-miR-148a and hsa-miR-424) in MSCs undergoing osteoblast differentiation have been predicted to target the mRNAs of Runx2, Cbfb, and BMPs; whereas hsa-miR-30c, hsa-miR-15b and hsa-miR-130b have been predicted to target MSC markers[141].
Table 1.
Non-exhaustive list of miRs that have been reported to inhibit or stimulate osteoblast differentiation in mouse or human
| miR | Species | Target | Effect | Ref. |
| MiR-206 | Mouse | Connexin 43 | Inhibitory | [129] |
| MiR-34 | Mouse | Satb 2 | Inhibitory | [130] |
| MiR-27a | Human | TNAP | Inhibitory | [131] |
| MiR-204 | Mouse | Runx 2 | Inhibitory | [132] |
| MiR-204/211 | Human | Runx 2 | Inhibitory | [143] |
| MiR-133 | Mouse | Runx 2 | Inhibitory | [133] |
| MiR-135 | Mouse | Smad 5 | Inhibitory | [133] |
| MiR-433 | Mouse | Runx 2 | Inhibitory | [134] |
| MiR-335 | Human | Runx 2 | Inhibitory | [142] |
| MiR-138 | Human | FAK | Inhibitory | [144] |
| MiR-2861 | Mouse | HDAC 5 | Stimulatory | [136] |
| MiR-335-5p | Mouse | Dkk 1 | Stimulatory | [137] |
| MiR-29a | Human | Dkk 1, Kremen 2 | Stimulatory | [138] |
MiRs: MicroRNAs; FAK: Focal adhesion kinase; HDAC 5: Histone deacetylase 5; TNAP: Tissue-nonspecific alkaline phosphatase; Dkk 1: Dickkopf-related protein 1; Runx 2: Runt-related 2; Satb2: Special AT-rich sequence binding protein 2; Smad5: Portmanteau of Sma in Drosophila and Mad in C. elegans.
Positive regulators: MiRs that may induce osteoblast differentiation include miR-2861, which promotes BMP2-induced ST2 osteoblast differentiation by repressing histone deacetylase 5 expression[142] (Table 1). MiR-335-5p also enhances osteogenic differentiation by inhibiting Dkk1 expression, and consequently by activating Wnt signaling[143]. Moreover, Kapinas et al[144] have shown that miR-29a promotes osteoblast differentiation by down regulating the inhibitors of canonical Wnt signaling such as Dkk1, Kremen2, and secreted frizzled related protein.
MiRs and hMSC
Several MiRs appear to significantly modulate osteoblast differentiation in mesenchymal precursors[145]. Dicer or Drosha knockdown in human MSCs inhibits osteogenic differentiation (reviewed in[146]). MiR expression patterns differ in MSC progenitors and fully differentiated cells, e.g., osteoblasts, adipocytes and chondrocytes suggesting that these miRs are important in MSC lineage decisions. Indeed, high or low expression of particular miRs may be a prerequisite for the commitment and differentiation of MSCs into specific lineages (reviewed in[147]). For instance, undifferentiated hMSCs isolated from various tissues were shown to express high levels of miR-335 while their differentiation resulted in a reduced expression of miR-335. The same miR as well as miR-204/211 impaired hMSC osteoblast differentiation by targeting Runx-2[148,149]. In human MSCs, decreased expression of miR-138 has also been associated with osteogenesis, possibly by targeting focal adhesion kinase[150]. Finally, and as presented above, MiR-148b, -27a, and -489 were found to play a critical role in early osteogenic differentiation of hMSC[137].
CONCLUSION
In the last decade, we have considerably increased our knowledge on the molecular contributors to osteoblast commitment and maturation. Since the discovery of the key role played by Runx2 in 1997[60], several other transcription factors have been demonstrated to modulate osteoblastogenesis. In addition, an increasing number of papers now indicate that the expression of these transcription factors is modulated by miRs, themselves being expressed under the control of the transcription factors they regulate[151]. Many of the results that had been obtained with murine models have now been confirmed with human MSCs. Collectively, the better understanding of the interaction between transcription factors and miRs, and of their effect on osteoblast to genesis and osteoblast function, will help develop new strategies to improve diagnosis and treatment of bone diseases.
Footnotes
P- Reviewers Goebel WS, Ma T, Kita K, Tamama K, Izeta A S- Editor Cui XM L- Editor A E- Editor Wang CH
References
- 1.Friedenstein AJ, Piatetzky-Shapiro II, Petrakova KV. Osteogenesis in transplants of bone marrow cells. J Embryol Exp Morphol. 1966;16:381–390. [PubMed] [Google Scholar]
- 2.Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S, Marshak DR. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284:143–147. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 3.Dominici M, Le Blanc K, Mueller I, Slaper-Cortenbach I, Marini F, Krause D, Deans R, Keating A, Prockop Dj, Horwitz E. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 2006;8:315–317. doi: 10.1080/14653240600855905. [DOI] [PubMed] [Google Scholar]
- 4.Lin CS, Xin ZC, Dai J, Lue TF. Commonly used mesenchymal stem cell markers and tracking labels: Limitations and challenges. Histol Histopathol. 2013;28:1109–1116. doi: 10.14670/hh-28.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caplan AI. Why are MSCs therapeutic? New data: new insight. J Pathol. 2009;217:318–324. doi: 10.1002/path.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zuk PA, Zhu M, Ashjian P, De Ugarte DA, Huang JI, Mizuno H, Alfonso ZC, Fraser JK, Benhaim P, Hedrick MH. Human adipose tissue is a source of multipotent stem cells. Mol Biol Cell. 2002;13:4279–4295. doi: 10.1091/mbc.E02-02-0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gronthos S, Mankani M, Brahim J, Robey PG, Shi S. Postnatal human dental pulp stem cells (DPSCs) in vitro and in vivo. Proc Natl Acad Sci USA. 2000;97:13625–13630. doi: 10.1073/pnas.240309797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mareschi K, Biasin E, Piacibello W, Aglietta M, Madon E, Fagioli F. Isolation of human mesenchymal stem cells: bone marrow versus umbilical cord blood. Haematologica. 2001;86:1099–1100. [PubMed] [Google Scholar]
- 9.Sacchetti B, Funari A, Michienzi S, Di Cesare S, Piersanti S, Saggio I, Tagliafico E, Ferrari S, Robey PG, Riminucci M, et al. Self-renewing osteoprogenitors in bone marrow sinusoids can organize a hematopoietic microenvironment. Cell. 2007;131:324–336. doi: 10.1016/j.cell.2007.08.025. [DOI] [PubMed] [Google Scholar]
- 10.Keating A. Mesenchymal stromal cells: new directions. Cell Stem Cell. 2012;10:709–716. doi: 10.1016/j.stem.2012.05.015. [DOI] [PubMed] [Google Scholar]
- 11.Ding J, Ghali O, Lencel P, Broux O, Chauveau C, Devedjian JC, Hardouin P, Magne D. TNF-alpha and IL-1beta inhibit RUNX2 and collagen expression but increase alkaline phosphatase activity and mineralization in human mesenchymal stem cells. Life Sci. 2009;84:499–504. doi: 10.1016/j.lfs.2009.01.013. [DOI] [PubMed] [Google Scholar]
- 12.Oshina H, Sotome S, Yoshii T, Torigoe I, Sugata Y, Maehara H, Marukawa E, Omura K, Shinomiya K. Effects of continuous dexamethasone treatment on differentiation capabilities of bone marrow-derived mesenchymal cells. Bone. 2007;41:575–583. doi: 10.1016/j.bone.2007.06.022. [DOI] [PubMed] [Google Scholar]
- 13.Murshed M, Harmey D, Millán JL, McKee MD, Karsenty G. Unique coexpression in osteoblasts of broadly expressed genes accounts for the spatial restriction of ECM mineralization to bone. Genes Dev. 2005;19:1093–1104. doi: 10.1101/gad.1276205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee NK, Sowa H, Hinoi E, Ferron M, Ahn JD, Confavreux C, Dacquin R, Mee PJ, McKee MD, Jung DY, et al. Endocrine regulation of energy metabolism by the skeleton. Cell. 2007;130:456–469. doi: 10.1016/j.cell.2007.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bonewald LF. The amazing osteocyte. J Bone Miner Res. 2011;26:229–238. doi: 10.1002/jbmr.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kon E, Filardo G, Roffi A, Di Martino A, Hamdan M, De Pasqual L, Merli ML, Marcacci M. Bone regeneration with mesenchymal stem cells. Clin Cases Miner Bone Metab. 2012;9:24–27. [PMC free article] [PubMed] [Google Scholar]
- 17.Marie PJ. Cell and gene therapy for bone repair. Osteoporos Int. 2011;22:2023–2026. doi: 10.1007/s00198-011-1615-0. [DOI] [PubMed] [Google Scholar]
- 18.Horwitz EM, Prockop DJ, Fitzpatrick LA, Koo WW, Gordon PL, Neel M, Sussman M, Orchard P, Marx JC, Pyeritz RE, et al. Transplantability and therapeutic effects of bone marrow-derived mesenchymal cells in children with osteogenesis imperfecta. Nat Med. 1999;5:309–313. doi: 10.1038/6529. [DOI] [PubMed] [Google Scholar]
- 19.Horwitz EM, Gordon PL, Koo WK, Marx JC, Neel MD, McNall RY, Muul L, Hofmann T. Isolated allogeneic bone marrow-derived mesenchymal cells engraft and stimulate growth in children with osteogenesis imperfecta: Implications for cell therapy of bone. Proc Natl Acad Sci USA. 2002;99:8932–8937. doi: 10.1073/pnas.132252399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Amini AR, Laurencin CT, Nukavarapu SP. Bone tissue engineering: recent advances and challenges. Crit Rev Biomed Eng. 2012;40:363–408. doi: 10.1615/critrevbiomedeng.v40.i5.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pochampally RR, Horwitz EM, DiGirolamo CM, Stokes DS, Prockop DJ. Correction of a mineralization defect by overexpression of a wild-type cDNA for COL1A1 in marrow stromal cells (MSCs) from a patient with osteogenesis imperfecta: a strategy for rescuing mutations that produce dominant-negative protein defects. Gene Ther. 2005;12:1119–1125. doi: 10.1038/sj.gt.3302514. [DOI] [PubMed] [Google Scholar]
- 22.Zhang R, Ducy P, Karsenty G. 1,25-dihydroxyvitamin D3 inhibits Osteocalcin expression in mouse through an indirect mechanism. J Biol Chem. 1997;272:110–116. doi: 10.1074/jbc.272.1.110. [DOI] [PubMed] [Google Scholar]
- 23.Clemens TL, Tang H, Maeda S, Kesterson RA, Demayo F, Pike JW, Gundberg CM. Analysis of osteocalcin expression in transgenic mice reveals a species difference in vitamin D regulation of mouse and human osteocalcin genes. J Bone Miner Res. 1997;12:1570–1576. doi: 10.1359/jbmr.1997.12.10.1570. [DOI] [PubMed] [Google Scholar]
- 24.Pinzone JJ, Hall BM, Thudi NK, Vonau M, Qiang YW, Rosol TJ, Shaughnessy JD. The role of Dickkopf-1 in bone development, homeostasis, and disease. Blood. 2009;113:517–525. doi: 10.1182/blood-2008-03-145169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Day TF, Guo X, Garrett-Beal L, Yang Y. Wnt/beta-catenin signaling in mesenchymal progenitors controls osteoblast and chondrocyte differentiation during vertebrate skeletogenesis. Dev Cell. 2005;8:739–750. doi: 10.1016/j.devcel.2005.03.016. [DOI] [PubMed] [Google Scholar]
- 26.Morvan F, Boulukos K, Clément-Lacroix P, Roman Roman S, Suc-Royer I, Vayssière B, Ammann P, Martin P, Pinho S, Pognonec P, et al. Deletion of a single allele of the Dkk1 gene leads to an increase in bone formation and bone mass. J Bone Miner Res. 2006;21:934–945. doi: 10.1359/jbmr.060311. [DOI] [PubMed] [Google Scholar]
- 27.Andrade AC, Nilsson O, Barnes KM, Baron J. Wnt gene expression in the post-natal growth plate: regulation with chondrocyte differentiation. Bone. 2007;40:1361–1369. doi: 10.1016/j.bone.2007.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bennett CN, Longo KA, Wright WS, Suva LJ, Lane TF, Hankenson KD, MacDougald OA. Regulation of osteoblastogenesis and bone mass by Wnt10b. Proc Natl Acad Sci USA. 2005;102:3324–3329. doi: 10.1073/pnas.0408742102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhong Z, Zylstra-Diegel CR, Schumacher CA, Baker JJ, Carpenter AC, Rao S, Yao W, Guan M, Helms JA, Lane NE, et al. Wntless functions in mature osteoblasts to regulate bone mass. Proc Natl Acad Sci USA. 2012;109:E2197–E2204. doi: 10.1073/pnas.1120407109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baksh D, Boland GM, Tuan RS. Cross-talk between Wnt signaling pathways in human mesenchymal stem cells leads to functional antagonism during osteogenic differentiation. J Cell Biochem. 2007;101:1109–1124. doi: 10.1002/jcb.21097. [DOI] [PubMed] [Google Scholar]
- 31.Boland GM, Perkins G, Hall DJ, Tuan RS. Wnt 3a promotes proliferation and suppresses osteogenic differentiation of adult human mesenchymal stem cells. J Cell Biochem. 2004;93:1210–1230. doi: 10.1002/jcb.20284. [DOI] [PubMed] [Google Scholar]
- 32.Zhou S. TGF-β regulates β-catenin signaling and osteoblast differentiation in human mesenchymal stem cells. J Cell Biochem. 2011;112:1651–1660. doi: 10.1002/jcb.23079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu G, Vijayakumar S, Grumolato L, Arroyave R, Qiao H, Akiri G, Aaronson SA. Canonical Wnts function as potent regulators of osteogenesis by human mesenchymal stem cells. J Cell Biol. 2009;185:67–75. doi: 10.1083/jcb.200810137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kang S, Bennett CN, Gerin I, Rapp LA, Hankenson KD, Macdougald OA. Wnt signaling stimulates osteoblastogenesis of mesenchymal precursors by suppressing CCAAT/enhancer-binding protein alpha and peroxisome proliferator-activated receptor gamma. J Biol Chem. 2007;282:14515–14524. doi: 10.1074/jbc.M700030200. [DOI] [PubMed] [Google Scholar]
- 35.Guo J, Jin J, Cooper LF. Dissection of sets of genes that control the character of wnt5a-deficient mouse calvarial cells. Bone. 2008;43:961–971. doi: 10.1016/j.bone.2008.06.011. [DOI] [PubMed] [Google Scholar]
- 36.Takada I, Mihara M, Suzawa M, Ohtake F, Kobayashi S, Igarashi M, Youn MY, Takeyama K, Nakamura T, Mezaki Y, et al. A histone lysine methyltransferase activated by non-canonical Wnt signalling suppresses PPAR-gamma transactivation. Nat Cell Biol. 2007;9:1273–1285. doi: 10.1038/ncb1647. [DOI] [PubMed] [Google Scholar]
- 37.Bilkovski R, Schulte DM, Oberhauser F, Gomolka M, Udelhoven M, Hettich MM, Roth B, Heidenreich A, Gutschow C, Krone W, et al. Role of WNT-5a in the determination of human mesenchymal stem cells into preadipocytes. J Biol Chem. 2010;285:6170–6178. doi: 10.1074/jbc.M109.054338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Briolay A, Lencel P, Bessueille L, Caverzasio J, Buchet R, Magne D. Autocrine stimulation of osteoblast activity by Wnt5a in response to TNF-α in human mesenchymal stem cells. Biochem Biophys Res Commun. 2013;430:1072–1077. doi: 10.1016/j.bbrc.2012.12.036. [DOI] [PubMed] [Google Scholar]
- 39.Chang J, Sonoyama W, Wang Z, Jin Q, Zhang C, Krebsbach PH, Giannobile W, Shi S, Wang CY. Noncanonical Wnt-4 signaling enhances bone regeneration of mesenchymal stem cells in craniofacial defects through activation of p38 MAPK. J Biol Chem. 2007;282:30938–30948. doi: 10.1074/jbc.M702391200. [DOI] [PubMed] [Google Scholar]
- 40.Li X, Cao X. BMP signaling and skeletogenesis. Ann N Y Acad Sci. 2006;1068:26–40. doi: 10.1196/annals.1346.006. [DOI] [PubMed] [Google Scholar]
- 41.Canalis E, Economides AN, Gazzerro E. Bone morphogenetic proteins, their antagonists, and the skeleton. Endocr Rev. 2003;24:218–235. doi: 10.1210/er.2002-0023. [DOI] [PubMed] [Google Scholar]
- 42.Urist MR. Bone: formation by autoinduction. Science. 1965;150:893–899. doi: 10.1126/science.150.3698.893. [DOI] [PubMed] [Google Scholar]
- 43.Urist MR, Strates BS. Bone morphogenetic protein. J Dent Res. 1971;50:1392–1406. doi: 10.1177/00220345710500060601. [DOI] [PubMed] [Google Scholar]
- 44.Massagué J. TGF-beta signal transduction. Annu Rev Biochem. 1998;67:753–791. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- 45.Guicheux J, Lemonnier J, Ghayor C, Suzuki A, Palmer G, Caverzasio J. Activation of p38 mitogen-activated protein kinase and c-Jun-NH2-terminal kinase by BMP-2 and their implication in the stimulation of osteoblastic cell differentiation. J Bone Miner Res. 2003;18:2060–2068. doi: 10.1359/jbmr.2003.18.11.2060. [DOI] [PubMed] [Google Scholar]
- 46.Lyons KM, Pelton RW, Hogan BL. Patterns of expression of murine Vgr-1 and BMP-2a RNA suggest that transforming growth factor-beta-like genes coordinately regulate aspects of embryonic development. Genes Dev. 1989;3:1657–1668. doi: 10.1101/gad.3.11.1657. [DOI] [PubMed] [Google Scholar]
- 47.Duprez D, Bell EJ, Richardson MK, Archer CW, Wolpert L, Brickell PM, Francis-West PH. Overexpression of BMP-2 and BMP-4 alters the size and shape of developing skeletal elements in the chick limb. Mech Dev. 1996;57:145–157. doi: 10.1016/0925-4773(96)00540-0. [DOI] [PubMed] [Google Scholar]
- 48.Zaidi M. Skeletal remodeling in health and disease. Nat Med. 2007;13:791–801. doi: 10.1038/nm1593. [DOI] [PubMed] [Google Scholar]
- 49.Yamaguchi A, Ishizuya T, Kintou N, Wada Y, Katagiri T, Wozney JM, Rosen V, Yoshiki S. Effects of BMP-2, BMP-4, and BMP-6 on osteoblastic differentiation of bone marrow-derived stromal cell lines, ST2 and MC3T3-G2/PA6. Biochem Biophys Res Commun. 1996;220:366–371. doi: 10.1006/bbrc.1996.0411. [DOI] [PubMed] [Google Scholar]
- 50.Bessa PC, Casal M, Reis RL. Bone morphogenetic proteins in tissue engineering: the road from laboratory to clinic, part II (BMP delivery) J Tissue Eng Regen Med. 2008;2:81–96. doi: 10.1002/term.74. [DOI] [PubMed] [Google Scholar]
- 51.Bessa PC, Casal M, Reis RL. Bone morphogenetic proteins in tissue engineering: the road from the laboratory to the clinic, part I (basic concepts) J Tissue Eng Regen Med. 2008;2:1–13. doi: 10.1002/term.63. [DOI] [PubMed] [Google Scholar]
- 52.Boerckel JD, Kolambkar YM, Dupont KM, Uhrig BA, Phelps EA, Stevens HY, García AJ, Guldberg RE. Effects of protein dose and delivery system on BMP-mediated bone regeneration. Biomaterials. 2011;32:5241–5251. doi: 10.1016/j.biomaterials.2011.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Agarwal R, Williams K, Umscheid CA, Welch WC. Osteoinductive bone graft substitutes for lumbar fusion: a systematic review. J Neurosurg Spine. 2009;11:729–740. doi: 10.3171/2009.6.SPINE08669. [DOI] [PubMed] [Google Scholar]
- 54.Garrison KR, Shemilt I, Donell S, Ryder JJ, Mugford M, Harvey I, Song F, Alt V. Bone morphogenetic protein (BMP) for fracture healing in adults. Cochrane Database Syst Rev. 2010;(6):CD006950. doi: 10.1002/14651858.CD006950.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Toth JM, Boden SD, Burkus JK, Badura JM, Peckham SM, McKay WF. Short-term osteoclastic activity induced by locally high concentrations of recombinant human bone morphogenetic protein-2 in a cancellous bone environment. Spine (Phila Pa 1976) 2009;34:539–550. doi: 10.1097/BRS.0b013e3181952695. [DOI] [PubMed] [Google Scholar]
- 56.Hill TP, Später D, Taketo MM, Birchmeier W, Hartmann C. Canonical Wnt/beta-catenin signaling prevents osteoblasts from differentiating into chondrocytes. Dev Cell. 2005;8:727–738. doi: 10.1016/j.devcel.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 57.Brault V, Moore R, Kutsch S, Ishibashi M, Rowitch DH, McMahon AP, Sommer L, Boussadia O, Kemler R. Inactivation of the beta-catenin gene by Wnt1-Cre-mediated deletion results in dramatic brain malformation and failure of craniofacial development. Development. 2001;128:1253–1264. doi: 10.1242/dev.128.8.1253. [DOI] [PubMed] [Google Scholar]
- 58.Gaur T, Lengner CJ, Hovhannisyan H, Bhat RA, Bodine PV, Komm BS, Javed A, van Wijnen AJ, Stein JL, Stein GS, et al. Canonical WNT signaling promotes osteogenesis by directly stimulating Runx2 gene expression. J Biol Chem. 2005;280:33132–33140. doi: 10.1074/jbc.M500608200. [DOI] [PubMed] [Google Scholar]
- 59.Komori T. Signaling networks in RUNX2-dependent bone development. J Cell Biochem. 2011;112:750–755. doi: 10.1002/jcb.22994. [DOI] [PubMed] [Google Scholar]
- 60.Ducy P, Zhang R, Geoffroy V, Ridall AL, Karsenty G. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell. 1997;89:747–754. doi: 10.1016/s0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]
- 61.Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson RT, Gao YH, Inada M, et al. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–764. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- 62.Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR, Stamp GW, Beddington RS, Mundlos S, Olsen BR, et al. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89:765–771. doi: 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- 63.Mundlos S, Otto F, Mundlos C, Mulliken JB, Aylsworth AS, Albright S, Lindhout D, Cole WG, Henn W, Knoll JH, et al. Mutations involving the transcription factor CBFA1 cause cleidocranial dysplasia. Cell. 1997;89:773–779. doi: 10.1016/s0092-8674(00)80260-3. [DOI] [PubMed] [Google Scholar]
- 64.Liu W, Toyosawa S, Furuichi T, Kanatani N, Yoshida C, Liu Y, Himeno M, Narai S, Yamaguchi A, Komori T. Overexpression of Cbfa1 in osteoblasts inhibits osteoblast maturation and causes osteopenia with multiple fractures. J Cell Biol. 2001;155:157–166. doi: 10.1083/jcb.200105052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kanatani N, Fujita T, Fukuyama R, Liu W, Yoshida CA, Moriishi T, Yamana K, Miyazaki T, Toyosawa S, Komori T. Cbf beta regulates Runx2 function isoform-dependently in postnatal bone development. Dev Biol. 2006;296:48–61. doi: 10.1016/j.ydbio.2006.03.039. [DOI] [PubMed] [Google Scholar]
- 66.Komori T. Regulation of osteoblast differentiation by transcription factors. J Cell Biochem. 2006;99:1233–1239. doi: 10.1002/jcb.20958. [DOI] [PubMed] [Google Scholar]
- 67.Zhao Z, Zhao M, Xiao G, Franceschi RT. Gene transfer of the Runx2 transcription factor enhances osteogenic activity of bone marrow stromal cells in vitro and in vivo. Mol Ther. 2005;12:247–253. doi: 10.1016/j.ymthe.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 68.Byers BA, Guldberg RE, Hutmacher DW, García AJ. Effects of Runx2 genetic engineering and in vitro maturation of tissue-engineered constructs on the repair of critical size bone defects. J Biomed Mater Res A. 2006;76:646–655. doi: 10.1002/jbm.a.30549. [DOI] [PubMed] [Google Scholar]
- 69.Lee JS, Lee JM, Im GI. Electroporation-mediated transfer of Runx2 and Osterix genes to enhance osteogenesis of adipose stem cells. Biomaterials. 2011;32:760–768. doi: 10.1016/j.biomaterials.2010.09.042. [DOI] [PubMed] [Google Scholar]
- 70.Kanai F, Marignani PA, Sarbassova D, Yagi R, Hall RA, Donowitz M, Hisaminato A, Fujiwara T, Ito Y, Cantley LC, et al. TAZ: a novel transcriptional co-activator regulated by interactions with 14-3-3 and PDZ domain proteins. EMBO J. 2000;19:6778–6791. doi: 10.1093/emboj/19.24.6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hong JH, Yaffe MB. TAZ: a beta-catenin-like molecule that regulates mesenchymal stem cell differentiation. Cell Cycle. 2006;5:176–179. doi: 10.4161/cc.5.2.2362. [DOI] [PubMed] [Google Scholar]
- 72.Hong JH, Hwang ES, McManus MT, Amsterdam A, Tian Y, Kalmukova R, Mueller E, Benjamin T, Spiegelman BM, Sharp PA, et al. TAZ, a transcriptional modulator of mesenchymal stem cell differentiation. Science. 2005;309:1074–1078. doi: 10.1126/science.1110955. [DOI] [PubMed] [Google Scholar]
- 73.Cho HH, Shin KK, Kim YJ, Song JS, Kim JM, Bae YC, Kim CD, Jung JS. NF-kappaB activation stimulates osteogenic differentiation of mesenchymal stem cells derived from human adipose tissue by increasing TAZ expression. J Cell Physiol. 2010;223:168–177. doi: 10.1002/jcp.22024. [DOI] [PubMed] [Google Scholar]
- 74.Jang EJ, Jeong H, Kang JO, Kim NJ, Kim MS, Choi SH, Yoo SE, Hong JH, Bae MA, Hwang ES. TM-25659 enhances osteogenic differentiation and suppresses adipogenic differentiation by modulating the transcriptional co-activator TAZ. Br J Pharmacol. 2012;165:1584–1594. doi: 10.1111/j.1476-5381.2011.01664.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.FitzPatrick DR, Carr IM, McLaren L, Leek JP, Wightman P, Williamson K, Gautier P, McGill N, Hayward C, Firth H, et al. Identification of SATB2 as the cleft palate gene on 2q32-q33. Hum Mol Genet. 2003;12:2491–2501. doi: 10.1093/hmg/ddg248. [DOI] [PubMed] [Google Scholar]
- 76.Dobreva G, Chahrour M, Dautzenberg M, Chirivella L, Kanzler B, Fariñas I, Karsenty G, Grosschedl R. SATB2 is a multifunctional determinant of craniofacial patterning and osteoblast differentiation. Cell. 2006;125:971–986. doi: 10.1016/j.cell.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 77.Zhang J, Tu Q, Grosschedl R, Kim MS, Griffin T, Drissi H, Yang P, Chen J. Roles of SATB2 in osteogenic differentiation and bone regeneration. Tissue Eng Part A. 2011;17:1767–1776. doi: 10.1089/ten.tea.2010.0503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 79.Ye JH, Xu YJ, Gao J, Yan SG, Zhao J, Tu Q, Zhang J, Duan XJ, Sommer CA, Mostoslavsky G, et al. Critical-size calvarial bone defects healing in a mouse model with silk scaffolds and SATB2-modified iPSCs. Biomaterials. 2011;32:5065–5076. doi: 10.1016/j.biomaterials.2011.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nakashima K, Zhou X, Kunkel G, Zhang Z, Deng JM, Behringer RR, de Crombrugghe B. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell. 2002;108:17–29. doi: 10.1016/s0092-8674(01)00622-5. [DOI] [PubMed] [Google Scholar]
- 81.Nishio Y, Dong Y, Paris M, O’Keefe RJ, Schwarz EM, Drissi H. Runx2-mediated regulation of the zinc finger Osterix/Sp7 gene. Gene. 2006;372:62–70. doi: 10.1016/j.gene.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 82.Tang W, Li Y, Osimiri L, Zhang C. Osteoblast-specific transcription factor Osterix (Osx) is an upstream regulator of Satb2 during bone formation. J Biol Chem. 2011;286:32995–33002. doi: 10.1074/jbc.M111.244236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tu Q, Valverde P, Li S, Zhang J, Yang P, Chen J. Osterix overexpression in mesenchymal stem cells stimulates healing of critical-sized defects in murine calvarial bone. Tissue Eng. 2007;13:2431–2440. doi: 10.1089/ten.2006.0406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang B, Huang S, Pan L, Jia S. Enhancement of bone formation by genetically engineered human umbilical cord-derived mesenchymal stem cells expressing osterix. Oral Surg Oral Med Oral Pathol Oral Radiol. 2013;116:e221–e229. doi: 10.1016/j.oooo.2011.12.024. [DOI] [PubMed] [Google Scholar]
- 85.Lee KS, Kim HJ, Li QL, Chi XZ, Ueta C, Komori T, Wozney JM, Kim EG, Choi JY, Ryoo HM, et al. Runx2 is a common target of transforming growth factor beta1 and bone morphogenetic protein 2, and cooperation between Runx2 and Smad5 induces osteoblast-specific gene expression in the pluripotent mesenchymal precursor cell line C2C12. Mol Cell Biol. 2000;20:8783–8792. doi: 10.1128/mcb.20.23.8783-8792.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang YW, Yasui N, Ito K, Huang G, Fujii M, Hanai J, Nogami H, Ochi T, Miyazono K, Ito Y. A RUNX2/PEBP2alpha A/CBFA1 mutation displaying impaired transactivation and Smad interaction in cleidocranial dysplasia. Proc Natl Acad Sci USA. 2000;97:10549–10554. doi: 10.1073/pnas.180309597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Phimphilai M, Zhao Z, Boules H, Roca H, Franceschi RT. BMP signaling is required for RUNX2-dependent induction of the osteoblast phenotype. J Bone Miner Res. 2006;21:637–646. doi: 10.1359/JBMR.060109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Javed A, Bae JS, Afzal F, Gutierrez S, Pratap J, Zaidi SK, Lou Y, van Wijnen AJ, Stein JL, Stein GS, et al. Structural coupling of Smad and Runx2 for execution of the BMP2 osteogenic signal. J Biol Chem. 2008;283:8412–8422. doi: 10.1074/jbc.M705578200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Javed A, Afzal F, Bae JS, Gutierrez S, Zaidi K, Pratap J, van Wijnen AJ, Stein JL, Stein GS, Lian JB. Specific residues of RUNX2 are obligatory for formation of BMP2-induced RUNX2-SMAD complex to promote osteoblast differentiation. Cells Tissues Organs. 2009;189:133–137. doi: 10.1159/000151719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sowa H, Kaji H, Hendy GN, Canaff L, Komori T, Sugimoto T, Chihara K. Menin is required for bone morphogenetic protein 2- and transforming growth factor beta-regulated osteoblastic differentiation through interaction with Smads and Runx2. J Biol Chem. 2004;279:40267–40275. doi: 10.1074/jbc.M401312200. [DOI] [PubMed] [Google Scholar]
- 91.Smink JJ, Leutz A. Instruction of mesenchymal cell fate by the transcription factor C/EBPβ. Gene. 2012;497:10–17. doi: 10.1016/j.gene.2012.01.043. [DOI] [PubMed] [Google Scholar]
- 92.Wu Z, Xie Y, Bucher NL, Farmer SR. Conditional ectopic expression of C/EBP beta in NIH-3T3 cells induces PPAR gamma and stimulates adipogenesis. Genes Dev. 1995;9:2350–2363. doi: 10.1101/gad.9.19.2350. [DOI] [PubMed] [Google Scholar]
- 93.Hata K, Nishimura R, Ueda M, Ikeda F, Matsubara T, Ichida F, Hisada K, Nokubi T, Yamaguchi A, Yoneda T. A CCAAT/enhancer binding protein beta isoform, liver-enriched inhibitory protein, regulates commitment of osteoblasts and adipocytes. Mol Cell Biol. 2005;25:1971–1979. doi: 10.1128/MCB.25.5.1971-1979.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Henriquez B, Hepp M, Merino P, Sepulveda H, van Wijnen AJ, Lian JB, Stein GS, Stein JL, Montecino M. C/EBPβ binds the P1 promoter of the Runx2 gene and up-regulates Runx2 transcription in osteoblastic cells. J Cell Physiol. 2011;226:3043–3052. doi: 10.1002/jcp.22652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gutierrez S, Javed A, Tennant DK, van Rees M, Montecino M, Stein GS, Stein JL, Lian JB. CCAAT/enhancer-binding proteins (C/EBP) beta and delta activate osteocalcin gene transcription and synergize with Runx2 at the C/EBP element to regulate bone-specific expression. J Biol Chem. 2002;277:1316–1323. doi: 10.1074/jbc.M106611200. [DOI] [PubMed] [Google Scholar]
- 96.Tominaga H, Maeda S, Hayashi M, Takeda S, Akira S, Komiya S, Nakamura T, Akiyama H, Imamura T. CCAAT/enhancer-binding protein beta promotes osteoblast differentiation by enhancing Runx2 activity with ATF4. Mol Biol Cell. 2008;19:5373–5386. doi: 10.1091/mbc.E08-03-0329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wiper-Bergeron N, St-Louis C, Lee JM. CCAAT/Enhancer binding protein beta abrogates retinoic acid-induced osteoblast differentiation via repression of Runx2 transcription. Mol Endocrinol. 2007;21:2124–2135. doi: 10.1210/me.2006-0452. [DOI] [PubMed] [Google Scholar]
- 98.Dingwall M, Marchildon F, Gunanayagam A, Louis CS, Wiper-Bergeron N. Retinoic acid-induced Smad3 expression is required for the induction of osteoblastogenesis of mesenchymal stem cells. Differentiation. 2011;82:57–65. doi: 10.1016/j.diff.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 99.Kveiborg M, Sabatakos G, Chiusaroli R, Wu M, Philbrick WM, Horne WC, Baron R. DeltaFosB induces osteosclerosis and decreases adipogenesis by two independent cell-autonomous mechanisms. Mol Cell Biol. 2004;24:2820–2830. doi: 10.1128/MCB.24.7.2820-2830.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Eferl R, Hoebertz A, Schilling AF, Rath M, Karreth F, Kenner L, Amling M, Wagner EF. The Fos-related antigen Fra-1 is an activator of bone matrix formation. EMBO J. 2004;23:2789–2799. doi: 10.1038/sj.emboj.7600282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kenner L, Hoebertz A, Beil FT, Keon N, Karreth F, Eferl R, Scheuch H, Szremska A, Amling M, Schorpp-Kistner M, et al. Mice lacking JunB are osteopenic due to cell-autonomous osteoblast and osteoclast defects. J Cell Biol. 2004;164:613–623. doi: 10.1083/jcb.200308155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yang X, Matsuda K, Bialek P, Jacquot S, Masuoka HC, Schinke T, Li L, Brancorsini S, Sassone-Corsi P, Townes TM, et al. ATF4 is a substrate of RSK2 and an essential regulator of osteoblast biology; implication for Coffin-Lowry Syndrome. Cell. 2004;117:387–398. doi: 10.1016/s0092-8674(04)00344-7. [DOI] [PubMed] [Google Scholar]
- 103.Xiao G, Jiang D, Ge C, Zhao Z, Lai Y, Boules H, Phimphilai M, Yang X, Karsenty G, Franceschi RT. Cooperative interactions between activating transcription factor 4 and Runx2/Cbfa1 stimulate osteoblast-specific osteocalcin gene expression. J Biol Chem. 2005;280:30689–30696. doi: 10.1074/jbc.M500750200. [DOI] [PubMed] [Google Scholar]
- 104.Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, Sadri N, Yun C, Popko B, Paules R, et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell. 2003;11:619–633. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- 105.Bruedigam C, Koedam M, Chiba H, Eijken M, van Leeuwen JP. Evidence for multiple peroxisome proliferator-activated receptor gamma transcripts in bone: fine-tuning by hormonal regulation and mRNA stability. FEBS Lett. 2008;582:1618–1624. doi: 10.1016/j.febslet.2008.04.012. [DOI] [PubMed] [Google Scholar]
- 106.Bruedigam C, Eijken M, Koedam M, van de Peppel J, Drabek K, Chiba H, van Leeuwen JP. A new concept underlying stem cell lineage skewing that explains the detrimental effects of thiazolidinediones on bone. Stem Cells. 2010;28:916–927. doi: 10.1002/stem.405. [DOI] [PubMed] [Google Scholar]
- 107.Akune T, Ohba S, Kamekura S, Yamaguchi M, Chung UI, Kubota N, Terauchi Y, Harada Y, Azuma Y, Nakamura K, et al. PPARgamma insufficiency enhances osteogenesis through osteoblast formation from bone marrow progenitors. J Clin Invest. 2004;113:846–855. doi: 10.1172/JCI19900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Jeon MJ, Kim JA, Kwon SH, Kim SW, Park KS, Park SW, Kim SY, Shin CS. Activation of peroxisome proliferator-activated receptor-gamma inhibits the Runx2-mediated transcription of osteocalcin in osteoblasts. J Biol Chem. 2003;278:23270–23277. doi: 10.1074/jbc.M211610200. [DOI] [PubMed] [Google Scholar]
- 109.Lencel P, Delplace S, Hardouin P, Magne D. TNF-α stimulates alkaline phosphatase and mineralization through PPARγ inhibition in human osteoblasts. Bone. 2011;48:242–249. doi: 10.1016/j.bone.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 110.Krause U, Harris S, Green A, Ylostalo J, Zeitouni S, Lee N, Gregory CA. Pharmaceutical modulation of canonical Wnt signaling in multipotent stromal cells for improved osteoinductive therapy. Proc Natl Acad Sci USA. 2010;107:4147–4152. doi: 10.1073/pnas.0914360107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Liu J, Wang H, Zuo Y, Farmer SR. Functional interaction between peroxisome proliferator-activated receptor gamma and beta-catenin. Mol Cell Biol. 2006;26:5827–5837. doi: 10.1128/MCB.00441-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lu D, Carson DA. Repression of beta-catenin signaling by PPAR gamma ligands. Eur J Pharmacol. 2010;636:198–202. doi: 10.1016/j.ejphar.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Rahman S, Czernik PJ, Lu Y, Lecka-Czernik B. β-catenin directly sequesters adipocytic and insulin sensitizing activities but not osteoblastic activity of PPARγ2 in marrow mesenchymal stem cells. PLoS One. 2012;7:e51746. doi: 10.1371/journal.pone.0051746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kang Q, Song WX, Luo Q, Tang N, Luo J, Luo X, Chen J, Bi Y, He BC, Park JK, et al. A comprehensive analysis of the dual roles of BMPs in regulating adipogenic and osteogenic differentiation of mesenchymal progenitor cells. Stem Cells Dev. 2009;18:545–559. doi: 10.1089/scd.2008.0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Jackson SM, Demer LL. Peroxisome proliferator-activated receptor activators modulate the osteoblastic maturation of MC3T3-E1 preosteoblasts. FEBS Lett. 2000;471:119–124. doi: 10.1016/s0014-5793(00)01372-7. [DOI] [PubMed] [Google Scholar]
- 116.Yu WH, Li FG, Chen XY, Li JT, Wu YH, Huang LH, Wang Z, Li P, Wang T, Lahn BT, et al. PPARγ suppression inhibits adipogenesis but does not promote osteogenesis of human mesenchymal stem cells. Int J Biochem Cell Biol. 2012;44:377–384. doi: 10.1016/j.biocel.2011.11.013. [DOI] [PubMed] [Google Scholar]
- 117.Alastalo TP, Li M, Perez Vde J, Pham D, Sawada H, Wang JK, Koskenvuo M, Wang L, Freeman BA, Chang HY, et al. Disruption of PPARγ/β-catenin-mediated regulation of apelin impairs BMP-induced mouse and human pulmonary arterial EC survival. J Clin Invest. 2011;121:3735–3746. doi: 10.1172/JCI43382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Monsalve FA, Pyarasani RD, Delgado-Lopez F, Moore-Carrasco R. Peroxisome proliferator-activated receptor targets for the treatment of metabolic diseases. Mediators Inflamm. 2013;2013:549627. doi: 10.1155/2013/549627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Matsui T, Yamagishi S. Osteogenic differentiation of mesenchymal stem cells is preserved by partial, but not full peroxisome proliferator-activated receptor-γ agonist. Int J Cardiol. 2011;146:109–110. doi: 10.1016/j.ijcard.2010.09.079. [DOI] [PubMed] [Google Scholar]
- 120.Bialek P, Kern B, Yang X, Schrock M, Sosic D, Hong N, Wu H, Yu K, Ornitz DM, Olson EN, et al. A twist code determines the onset of osteoblast differentiation. Dev Cell. 2004;6:423–435. doi: 10.1016/s1534-5807(04)00058-9. [DOI] [PubMed] [Google Scholar]
- 121.el Ghouzzi V, Le Merrer M, Perrin-Schmitt F, Lajeunie E, Benit P, Renier D, Bourgeois P, Bolcato-Bellemin AL, Munnich A, Bonaventure J. Mutations of the TWIST gene in the Saethre-Chotzen syndrome. Nat Genet. 1997;15:42–46. doi: 10.1038/ng0197-42. [DOI] [PubMed] [Google Scholar]
- 122.Howard TD, Paznekas WA, Green ED, Chiang LC, Ma N, Ortiz de Luna RI, Garcia Delgado C, Gonzalez-Ramos M, Kline AD, Jabs EW. Mutations in TWIST, a basic helix-loop-helix transcription factor, in Saethre-Chotzen syndrome. Nat Genet. 1997;15:36–41. doi: 10.1038/ng0197-36. [DOI] [PubMed] [Google Scholar]
- 123.Danciu TE, Li Y, Koh A, Xiao G, McCauley LK, Franceschi RT. The basic helix loop helix transcription factor Twist1 is a novel regulator of ATF4 in osteoblasts. J Cell Biochem. 2012;113:70–79. doi: 10.1002/jcb.23329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Miraoui H, Severe N, Vaudin P, Pagès JC, Marie PJ. Molecular silencing of Twist1 enhances osteogenic differentiation of murine mesenchymal stem cells: implication of FGFR2 signaling. J Cell Biochem. 2010;110:1147–1154. doi: 10.1002/jcb.22628. [DOI] [PubMed] [Google Scholar]
- 125.Isenmann S, Arthur A, Zannettino AC, Turner JL, Shi S, Glackin CA, Gronthos S. TWIST family of basic helix-loop-helix transcription factors mediate human mesenchymal stem cell growth and commitment. Stem Cells. 2009;27:2457–2468. doi: 10.1002/stem.181. [DOI] [PubMed] [Google Scholar]
- 126.Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mourelatos Z. Small RNAs: The seeds of silence. Nature. 2008;455:44–45. doi: 10.1038/455044a. [DOI] [PubMed] [Google Scholar]
- 128.Wienholds E, Plasterk RH. MicroRNA function in animal development. FEBS Lett. 2005;579:5911–5922. doi: 10.1016/j.febslet.2005.07.070. [DOI] [PubMed] [Google Scholar]
- 129.Vickers KC, Remaley AT. MicroRNAs in atherosclerosis and lipoprotein metabolism. Curr Opin Endocrinol Diabetes Obes. 2010;17:150–155. doi: 10.1097/MED.0b013e32833727a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Dong S, Yang B, Guo H, Kang F. MicroRNAs regulate osteogenesis and chondrogenesis. Biochem Biophys Res Commun. 2012;418:587–591. doi: 10.1016/j.bbrc.2012.01.075. [DOI] [PubMed] [Google Scholar]
- 131.Kim EY, Moudgil KD. Regulation of autoimmune inflammation by pro-inflammatory cytokines. Immunol Lett. 2008;120:1–5. doi: 10.1016/j.imlet.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lindsay MA. microRNAs and the immune response. Trends Immunol. 2008;29:343–351. doi: 10.1016/j.it.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 133.Hassan MQ, Maeda Y, Taipaleenmaki H, Zhang W, Jafferji M, Gordon JA, Li Z, Croce CM, van Wijnen AJ, Stein JL, et al. miR-218 directs a Wnt signaling circuit to promote differentiation of osteoblasts and osteomimicry of metastatic cancer cells. J Biol Chem. 2012;287:42084–42092. doi: 10.1074/jbc.M112.377515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bianco P, Riminucci M, Gronthos S, Robey PG. Bone marrow stromal stem cells: nature, biology, and potential applications. Stem Cells. 2001;19:180–192. doi: 10.1634/stemcells.19-3-180. [DOI] [PubMed] [Google Scholar]
- 135.Inose H, Ochi H, Kimura A, Fujita K, Xu R, Sato S, Iwasaki M, Sunamura S, Takeuchi Y, Fukumoto S, et al. A microRNA regulatory mechanism of osteoblast differentiation. Proc Natl Acad Sci USA. 2009;106:20794–20799. doi: 10.1073/pnas.0909311106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wei J, Shi Y, Zheng L, Zhou B, Inose H, Wang J, Guo XE, Grosschedl R, Karsenty G. miR-34s inhibit osteoblast proliferation and differentiation in the mouse by targeting SATB2. J Cell Biol. 2012;197:509–521. doi: 10.1083/jcb.201201057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Schoolmeesters A, Eklund T, Leake D, Vermeulen A, Smith Q, Force Aldred S, Fedorov Y. Functional profiling reveals critical role for miRNA in differentiation of human mesenchymal stem cells. PLoS One. 2009;4:e5605. doi: 10.1371/journal.pone.0005605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Huang J, Zhao L, Xing L, Chen D. MicroRNA-204 regulates Runx2 protein expression and mesenchymal progenitor cell differentiation. Stem Cells. 2010;28:357–364. doi: 10.1002/stem.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Li Z, Hassan MQ, Volinia S, van Wijnen AJ, Stein JL, Croce CM, Lian JB, Stein GS. A microRNA signature for a BMP2-induced osteoblast lineage commitment program. Proc Natl Acad Sci USA. 2008;105:13906–13911. doi: 10.1073/pnas.0804438105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kim EJ, Kang IH, Lee JW, Jang WG, Koh JT. MiR-433 mediates ERRγ-suppressed osteoblast differentiation via direct targeting to Runx2 mRNA in C3H10T1/2 cells. Life Sci. 2013;92:562–568. doi: 10.1016/j.lfs.2013.01.015. [DOI] [PubMed] [Google Scholar]
- 141.Gao J, Yang T, Han J, Yan K, Qiu X, Zhou Y, Fan Q, Ma B. MicroRNA expression during osteogenic differentiation of human multipotent mesenchymal stromal cells from bone marrow. J Cell Biochem. 2011;112:1844–1856. doi: 10.1002/jcb.23106. [DOI] [PubMed] [Google Scholar]
- 142.Li H, Xie H, Liu W, Hu R, Huang B, Tan YF, Xu K, Sheng ZF, Zhou HD, Wu XP, et al. A novel microRNA targeting HDAC5 regulates osteoblast differentiation in mice and contributes to primary osteoporosis in humans. J Clin Invest. 2009;119:3666–3677. doi: 10.1172/JCI39832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zhang J, Tu Q, Bonewald LF, He X, Stein G, Lian J, Chen J. Effects of miR-335-5p in modulating osteogenic differentiation by specifically downregulating Wnt antagonist DKK1. J Bone Miner Res. 2011;26:1953–1963. doi: 10.1002/jbmr.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kapinas K, Kessler C, Ricks T, Gronowicz G, Delany AM. miR-29 modulates Wnt signaling in human osteoblasts through a positive feedback loop. J Biol Chem. 2010;285:25221–25231. doi: 10.1074/jbc.M110.116137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Taipaleenmäki H, Bjerre Hokland L, Chen L, Kauppinen S, Kassem M. Mechanisms in endocrinology: micro-RNAs: targets for enhancing osteoblast differentiation and bone formation. Eur J Endocrinol. 2012;166:359–371. doi: 10.1530/EJE-11-0646. [DOI] [PubMed] [Google Scholar]
- 146.Kapinas K, Delany AM. MicroRNA biogenesis and regulation of bone remodeling. Arthritis Res Ther. 2011;13:220. doi: 10.1186/ar3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Guo L, Zhao RC, Wu Y. The role of microRNAs in self-renewal and differentiation of mesenchymal stem cells. Exp Hematol. 2011;39:608–616. doi: 10.1016/j.exphem.2011.01.011. [DOI] [PubMed] [Google Scholar]
- 148.Tomé M, López-Romero P, Albo C, Sepúlveda JC, Fernández-Gutiérrez B, Dopazo A, Bernad A, González MA. miR-335 orchestrates cell proliferation, migration and differentiation in human mesenchymal stem cells. Cell Death Differ. 2011;18:985–995. doi: 10.1038/cdd.2010.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zhang Y, Xie RL, Croce CM, Stein JL, Lian JB, van Wijnen AJ, Stein GS. A program of microRNAs controls osteogenic lineage progression by targeting transcription factor Runx2. Proc Natl Acad Sci USA. 2011;108:9863–9868. doi: 10.1073/pnas.1018493108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Eskildsen T, Taipaleenmäki H, Stenvang J, Abdallah BM, Ditzel N, Nossent AY, Bak M, Kauppinen S, Kassem M. MicroRNA-138 regulates osteogenic differentiation of human stromal (mesenchymal) stem cells in vivo. Proc Natl Acad Sci USA. 2011;108:6139–6144. doi: 10.1073/pnas.1016758108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lian JB, Stein GS, van Wijnen AJ, Stein JL, Hassan MQ, Gaur T, Zhang Y. MicroRNA control of bone formation and homeostasis. Nat Rev Endocrinol. 2012;8:212–227. doi: 10.1038/nrendo.2011.234. [DOI] [PMC free article] [PubMed] [Google Scholar]


