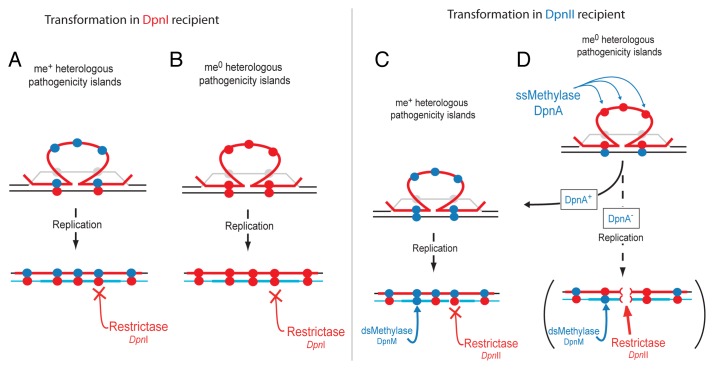
Figure 2. Differential impact of DpnI and DpnII R–M systems on transformation. (A) DpnI does not interfere with transformation of a pathogenicity island on me+ (closed blue circles) DNA. Transforming me+ ssDNA (red line) pairing with homologous DNA on host chromosome (black line) displaces the complementary strand (gray line). The pathogenicity island sequence remains in the form of ssDNA due to lack of homology. After replication, me+/0 sites are produced by synthesis of complementary neosynthesized DNA (light blue), which are not sensitive to restriction by the DpnI restrictase. (B) DpnI does not interfere with transformation of a pathogenicity island on me0 (closed red circles) DNA. DNA identity as in (A). (C) DpnII does not interfere with transformation of a pathogenicity island on me+ DNA. DNA identity as in (A). After replication, dsDNA is either fully me+ or me+/0 depending on the presence and action of methylases, and therefore resistant to DpnII. (D) Transformation of a pathogenicity island on me0 (closed red circles) transforming DNA into a DpnII strain in the presence or absence of ssDNA methylase DpnA. DNA identity as in (A). In the presence of DpnA, the ssDNA of the pathogenicity island in the transformation heteroduplex is methylated, and thus protected from DpnII restriction after replication, as in (C) (DpnA+ arrow). In the unnatural absence of DpnA, the ssDNA of the pathogenicity island remains me0 in the transformation heteroduplex. After replication (DpnA− arrow and brackets), me0 sites in the heterologous transforming DNA are paired with me0 sites in the neosynthesized DNA. Resulting dsDNA me0 sites in the chromosome are sensitive to DpnII, which can restrict the chromosome (red brackets in DNA) and kill the cell.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.