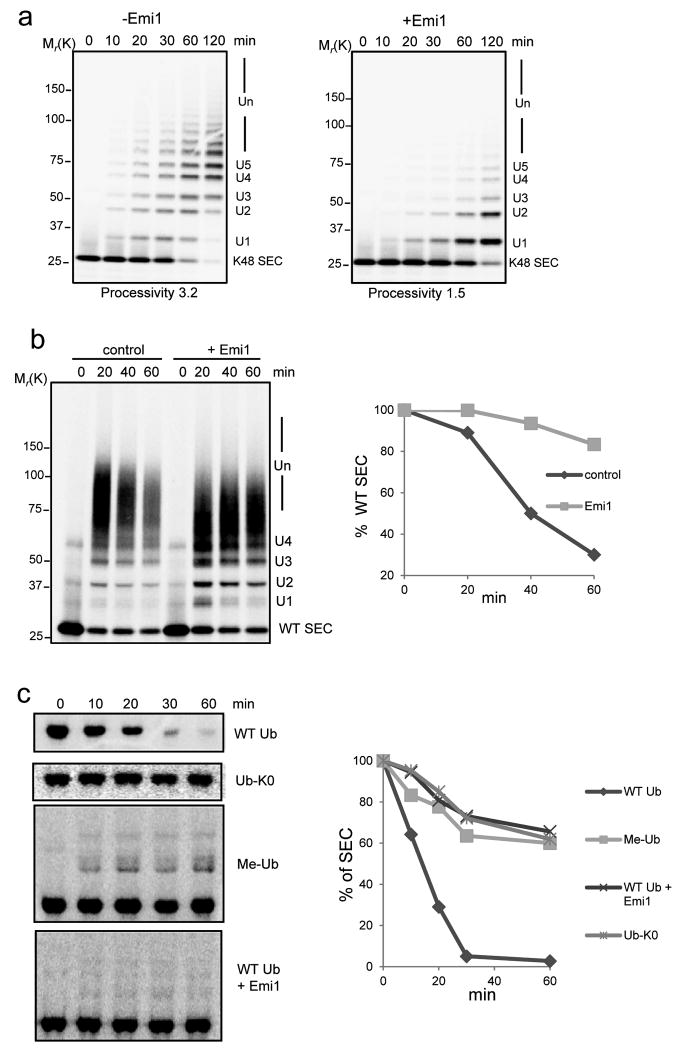
Figure 1.
Emi1 stabilizes substrates by preventing ubiquitin chain assembly.
(a) 200 nM 33P labeled single-lysine securin (K48 SEC) was ubiquitylated by 4 nM APCCdh1 in the presence (+) or absence (-) of 200 nM Emi1. The reactions were supplemented with UBCH10, Ube2S and WT ubiquitin, and analyzed by autoradiography. (b) Ubiquitylation and degradation of WT securin was examined in the presence or absence of Emi1. 100 nM 33P labeled WT securin, 4 nM purified APCCdh1, 100 nM Emi1 and 8 nM 26S proteasomes were used in the reconstituted reaction, supplemented with UBCH10, Ube2S and WT ubiquitin and analyzed by autoradiography. In the graphs the remaining substrates were quantified and plotted as percentages of input radiolabeled securin at indicated time points. These data are representative of three independent experiments. (c) Degradation of 35S labeled WT securin was examined in cell extracts prepared from HeLa S3 cells synchronized at G1 phase. WT ubiquitin (WT Ub), lysine-free ubiquitin (Ub-K0), methylated ubiquitin (Me-Ub) or Emi1 were added as indicated and analyzed by autoradiography. The amount of remaining securin was quantified and plotted at indicated time points. These data are representative of three independent experiments.