Figure 1. Cellular sites of membrane curvature.
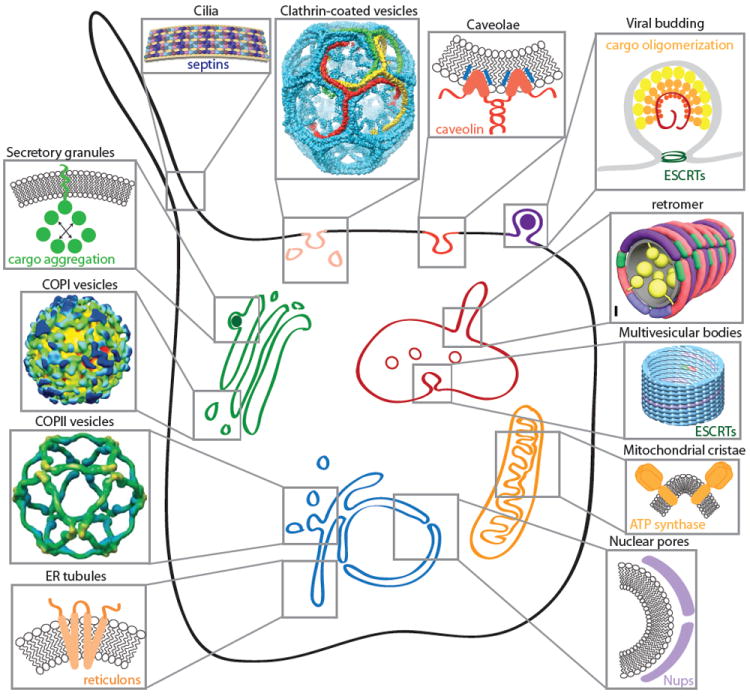
The membranes of eukaryotic cells display many instances of membrane curvature, some of which are dynamic (e.g. transport vesicles, endosomal tubules, viral buds) and others more static (e.g. nuclear pores, cilia, ER tubules, mitochondrial cristae). Each of these examples of membrane curvature is created by physical effects that derive from both lipid and protein sources. Self-assembling proteins can scaffold membranes (clathrin, COPI, COPII, nucleoporins, caveolins, reticulons, retromer, ESCRTs and septins). Asymmetric lipid and protein insertion can drive curvature by the bilayer couple model and molecular crowding effects (secretory granule cargoes, reticulons, caveolins, viral matrix proteins, mitochondrial ATP synthase). COPI structure reprinted from Faini et al. 46, copyright 2004, with permission from AAAS. COPII structure reprinted from Stagg et al 45, clathrin structure reprinted from Fotin et al 83, and retromer model reprinted from Hierro et al 84. Septin model reprinted from Tanaka-Takiguchi et al. 85, copyright 2009, with permission from Elsevier. ESCRT structure reprinted from Effantin et al.86, copyright 2013, with permission from Wiley.
