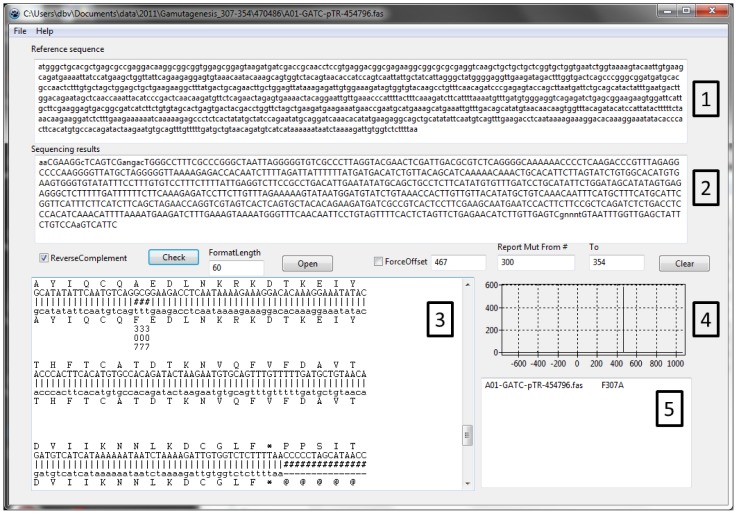
Figure 3. Mutant Checker interface.
(1) Input text box for reference sequence, starting with the beginning of the expressed sequence (first ATG). (2) Input text box for sequencing results. The sequence is reverse completented for the analyis if the “ReverseComplement” checkbox is activated. (3) Output windows of aligned sequences, DNA to protein translation of both reference sequence showing identified mutations and their position, based on the reference sequence. (4) Graphical output of the aligment score function vs offset between to sequences. A single peak indicate only one possible aligment, while multiple peaks indicate several alternative aligments and non-productive clone. (5) Identified mutations, within the specified region.