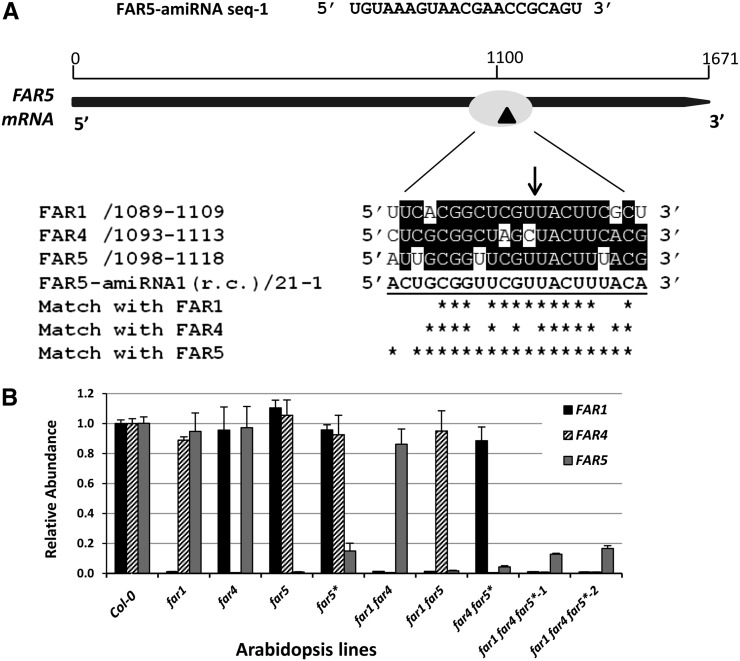
Figure 2.
Generation of double and triple far mutant lines by amiRNA silencing. A, Schematic showing the FAR5-amiRNA-sequence 1 and the cleavage site on FAR5 cDNA sequence. The reverse complement (r.c.) FAR5-amiRNA-sequence 1 aligned with the coding sequence of root-expressing FARs FAR1, FAR4, and FAR5. Ideal amiRNA sequences have no mismatches in positions 2 and 12 and only have one or two mismatches in positions 18 to 21 (Ossowski et al., 2008). Cross hybridization of FAR5-amiRNA-sequence 1 with FAR1 and FAR4 is unlikely because of several mismatches in the 5′ end region. B, Transcript levels of FAR genes in wild-type, far1, far4, far5, far double, and far triple mutant lines determined by quantitative RT-PCR (*, FAR5 amiRNA lines; 1 and 2, two independent transgenic far triple mutant lines). Total RNA (0.1 µg) isolated from the roots of 2-week-old T5 seedlings grown on MS agar plates was used for quantitative RT-PCR. Results are presented as relative transcript abundances to Col-0 and normalized through geometric averaging of three constitutively expressed genes (ACT2, GAPC, and eIF4a1). The data represent means of two biological replicates, each with three technical replicates.