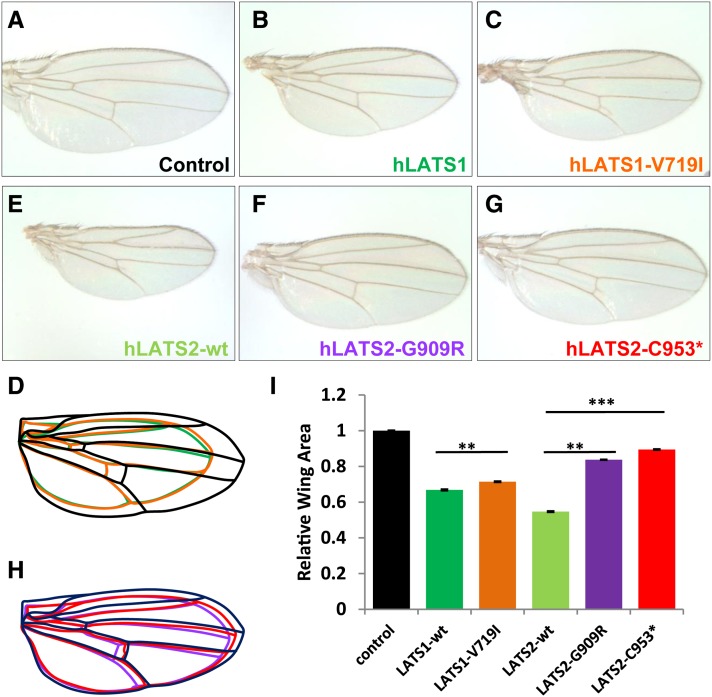
Figure 1.
In vivo analysis of hLATS1/2 and variants for their activity to inhibit tissue growth in Drosophila. (A–H) Images of female wings (anterior to the top) that express following transgenes under the control of MS1096-Gal4. (A) UAS-86Fb-lacZ as a negative control. (B) UAS-86Fb-hLATS1-wt. (C) UAS-86Fb-hLATS1-V719I. (D) Outlines of the wings shown in A–C. (E) UAS-86Fb-hLATS2-wt. (F) UAS-86Fb-hLATS2-G909R. (G) UAS-86Fb-hLATS2-C953*. (H) Outlines of the wings shown in E–G. (I) Quantification of total wing area of the genotypes (n = 40 for each genotype) displayed in A–C and E–G. Results represent mean ± SEM. **P < 10−11, ***P < 10−53. Embryos from Drosophila attP docking strains y[1] M{vas-int.Dm}ZH-2A w[*]; M{3xP3-RFP.attP}ZH-86Fb or y[1] M{vas-int.Dm}ZH-2A w[*]; M{3xP3-RFP.attP}ZH-51D (from Bloomington Drosophila Stock Center, BDSC; Bischof et al. 2007) were injected with the pUAST-attB constructs carrying wild-type hLATS1/2 and mutant variants. Fly embryo injections were partly done by Rainbow Transgenic Flies. Orange eye color was a selection marker for isolating transformants. Transgenic lines were established through standard procedures. Flies with pUAST-lacZ-attB inserted in 51D or 86Fb (gift from Johannes Bischof) were used as controls. The Drosophila strains that carry wts-RNAi on the second chromosome (no. v106174) and the third chromosome (no. 34064) were obtained from the Vienna Drosophila RNAi Center (VDRC) and BDSC, respectively. The VDRC UAS-wts-RNAi inserted in the second chromosome was combined with UAS-hLATS1/2 in the third chromosome (86Fb), while the BDSC UAS-wts-RNAi inserted in the third chromosome was combined with UAS-hLATS1/2 at the second chromosome (51D). MS1096-Gal4 was used to drive wing-specific expression. Adult wings were dissected and analyzed by ImageJ for size measurement.