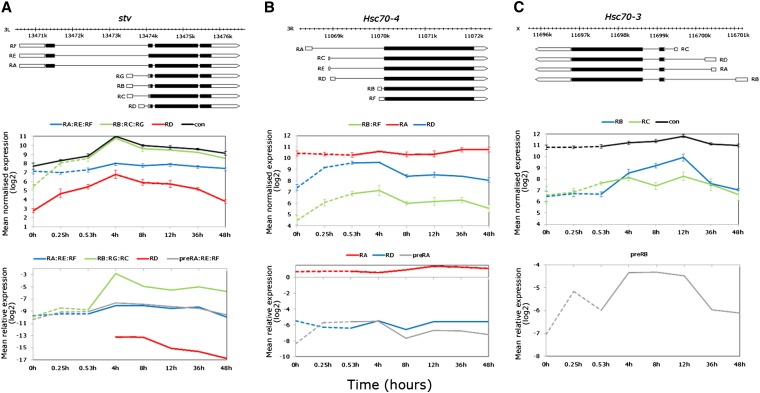
Figure 8.
Elucidating expression complexity of alternatively transcribed genes upregulated during thermal stress in the exon data. (A) stv: gene model showing the long and short transcript subsets derived by alternative transcription and alternative splicing (top); average normalized log2 exon expression for the RA:RE:RF, RB:RC:RG subsets and RD and constitutive exons (middle); and real-time PCR data relative to RpL11 showing no expression variation of the RA:RE:RF subset, accumulation of the primary transcript during heat stress, and induction of the RB:RC:RG subset and RD isoform by alternative transcription only during recovery. (B) Hsc70-4: gene model showing isoforms derived by alternative transcription (top); average normalized log2 exon expression of the RA, and RD isoforms, and RB:RF subset (middle); and real-time PCR data showing only accumulation of the primary transcript during heat stress (bottom). (C) Hsc70-3: gene model showing isoforms derived by alternative transcription (top); average normalized log2 exon expression of long (RB, recovery induced), and short (RC, stress induced) (middle); and real time PCR data for only the primary transcript showing accumulation during heat stress (bottom). The PCR data were log2 transformed for visual clarity and nontransformed data with error bars (±SE of the mean) are shown in Table S7.