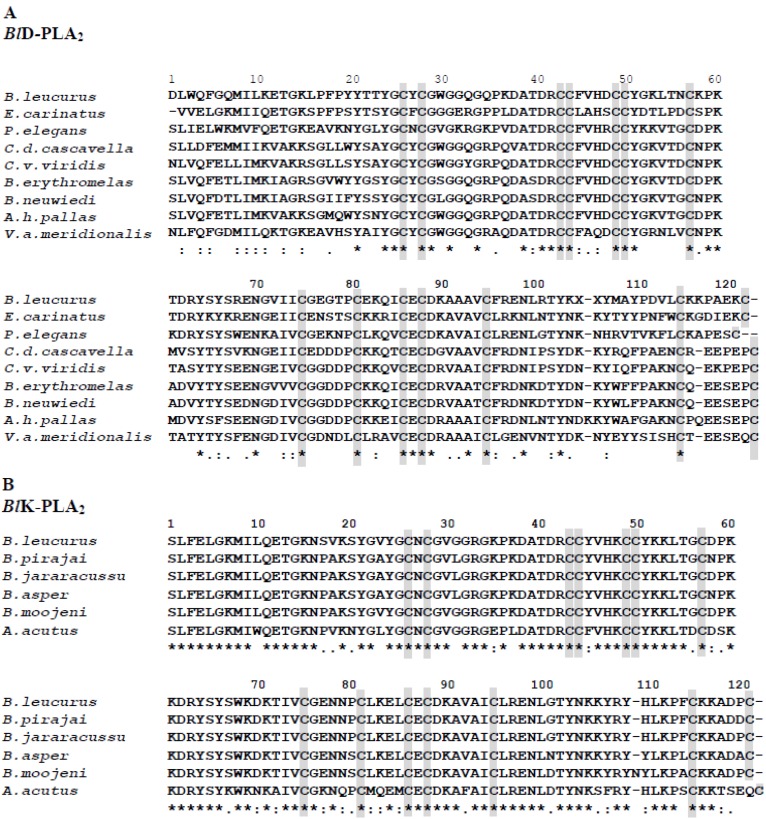
Figure 2.
Multiple amino acid sequence alignment of BlPLA2s with svPLA2s homologous. The one letter code for amino acid nomenclature is used. Proteins compared and their UniProt or GeneBank (GB) accession numbers: K49 (piratoxin II, P82287) from Bothrops pirajai: K49 (bthtx I, Q90249) from B. jararacussu: K49 (BlK-PLA2, P86975) from B. leucurus: K49 (myotoxin II, P24605) from B. asper: K49 (myotoxin II, Q91834) from B. moojeni: K49 (O57385) from Deinagkistrodon (formerly Agkistrodon) acutus: R49 (Q28681) from Protobothrops elegans: S49 (P48650) from Echis carinatus; D49 (BlD-PLA2, P86974) from B. leucurus: D49 (vipoxin complex, P04084) from Vipera ammodytes meridionalis: D49 (C9E7C4) from Crotalus durissus cascavella: D49 (Q7ZTA6) from C.v.viridis: D49 (Q2H228) from B. erythromelas: D49 (GB/KC544002) from B. neuwiedi: D49 (O42191) from Gloydius (formerly Agkistrodon) halys pallas. (*) is used for identical residues (:) for conserved ones and (.) for semi-conserved substitutions among all sequences in the alignment. Gaps were introduced to maximize the sequence homology, as indicated in part (A) and (B), respectively.