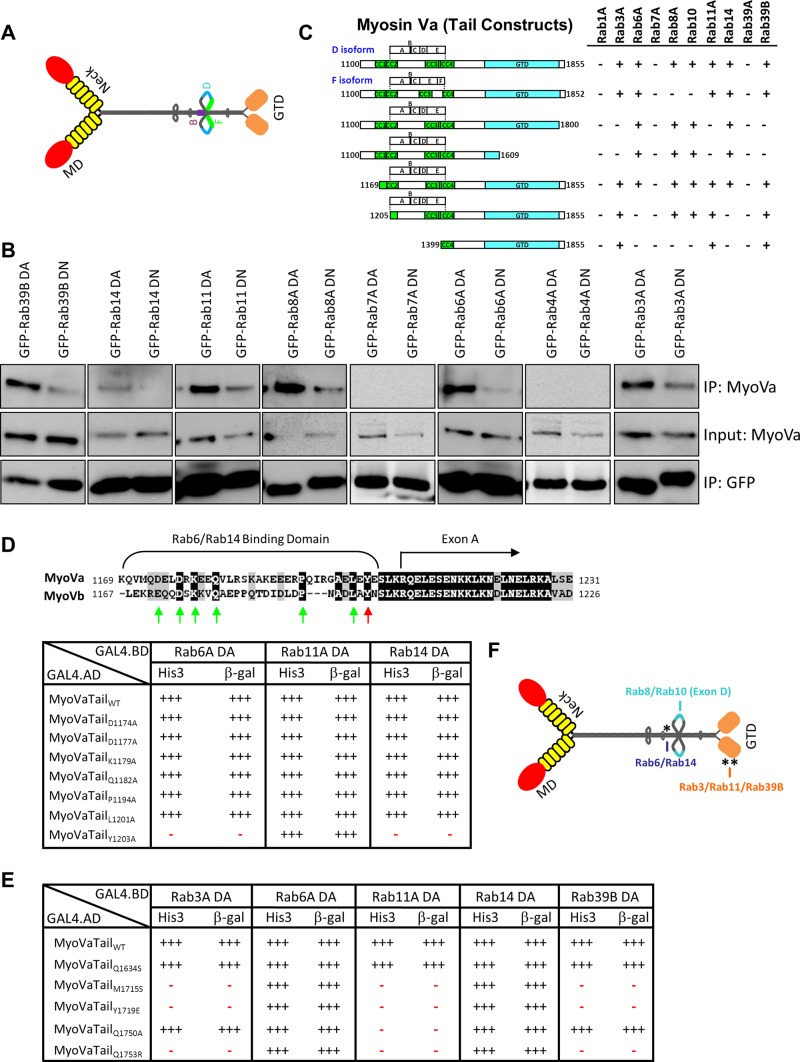
FIGURE 1:
Myosin Va interacts with multiple Rab GTPases. (A) Schematic diagram depicting the extended “active” structure of myosin Va. The location of the alternatively spliced exons B, D, and F are indicated. GTD, globular-tail domain; MD, motor domain. (B) HeLa cells expressing dominant-active (DA) or dominant-negative (DN) mutants of the indicated Rab GTPases fused to GFP were lysed, and the fusion proteins were immunoprecipitated with an anti-GFP antibody. The ability of myosin Va to form a complex with these Rabs was tested by Western blot analysis using anti-myosin Va antibody. Input represents 10% of the starting material used in all conditions. (C) Regions of the myosin Va tail tested using the yeast two-hybrid technique. The constitutively active mutant of each Rab GTPase was used. CC, coiled coil; GTD, globular tail domain. (D) Identification of a critical amino acid for binding to Rab6 and Rab14. ClustalW alignment of the Rab6/Rab14-binding regions of human myosin Va and myosin Vb. Arrows indicate the conserved amino acids that were mutated to alanine, and the red arrow indicates tyrosine 1203, which, when mutated, abolishes the Rab6 and Rab14 interaction. The table indicates the strength of interaction of each mutant with constitutively active Rab6A, Rab11A, and Rab14 using the yeast two-hybrid HIS3 and LacZ reporter assays. (E) Identification of critical amino acids that mediate binding to Rab3A, Rab11A, and Rab39B. The amino acids in myosin Va that correspond to those identified by Lipatova et al. (2008) that mediate the binding of Myo2 to Ypt31/32 were mutated and the mutants tested for interaction with the constitutively active mutants of the indicated Rab GTPases by yeast two-hybrid assay. (F) Schematic diagram of myosin Va indicating the identified Rab-binding domains. Asterisk indicates the location of the Y1203A mutation; double asterisks indicate the location of the Q1753R mutation.