FIGURE 3:
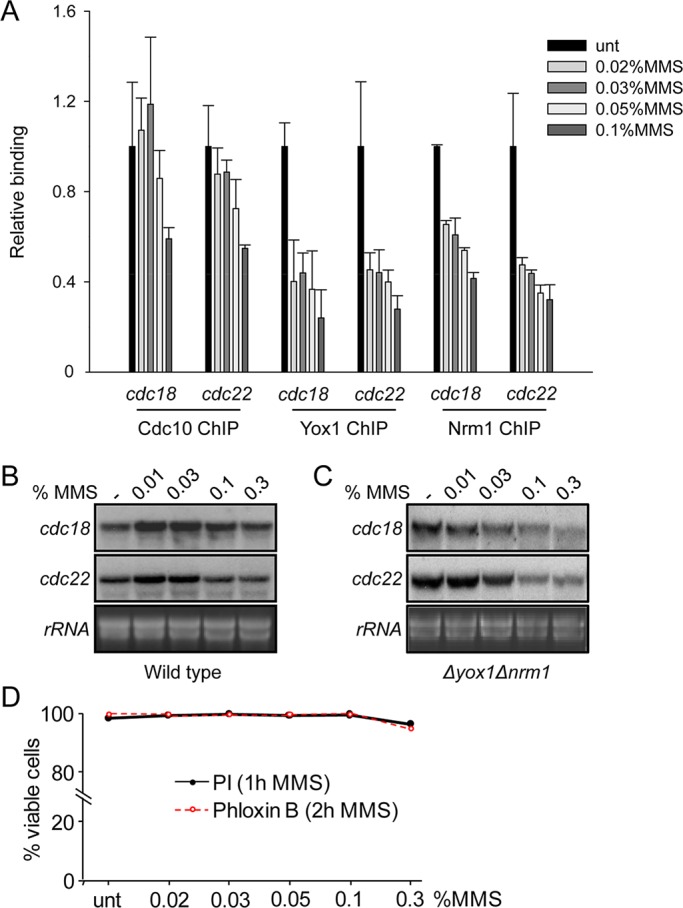
Chk1 effect on Cdc10 depends on MMS concentration. (A) Loading of Cdc10, Yox1, and Nrm1-HA on cdc22 and cdc18 promoters was measured by ChIP analysis of chromatin extracts isolated from untreated or MMS-treated cultures with the indicated concentrations (1 h at 30ºC). Cdc10 and Nrm1 are HA tagged, and the levels of binding are quantified on anti-HA immunoprecipitated DNA, whereas Yox1 is determined with anti-Yox1polyclonal antibodies. (B) Total RNA was prepared from untreated (–) or MMS-treated cultures of wild-type cells and analyzed by hybridization to the probes indicated on the left. rRNA is shown as loading control. (C) Total RNA was prepared from untreated or MMS-treated (increasing doses) cultures of Δyox1Δnrm1 cells and analyzed by hybridization with the probes indicated on the left. rRNA is shown as loading control. (D) Cell viability is unaffected at the used range of MMS concentrations. Viability test of wild-type cells treated with different concentrations of MMS, using propidium iodide or phloxine to measure viable cells, was performed by fluorescence-activated cell sorting analysis.
