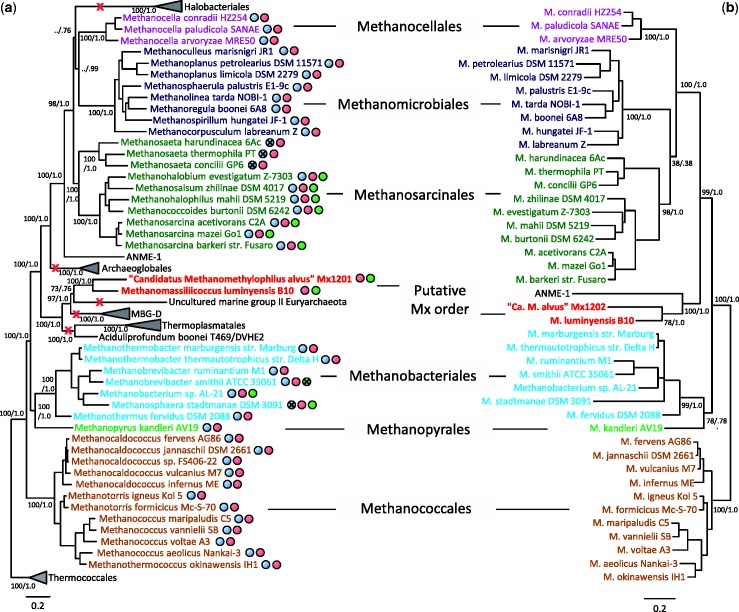
Fig. 2.—
Phylogenetic position of the Mx order and inferred losses of methanogenesis. (a) Bayesian phylogeny of Euryarchaeota based on a concatenation of 57 ribosomal proteins comprising 7,472 amino acid positions. Values at nodes represent Bayesian posterior probabilities and bootstrap values based on maximum likelihood analysis and 100 resamplings of the original data set. For clarity, only the values corresponding to the monophyly of orders and their evolutionary relationships are shown. “../” symbols indicate that the corresponding node was not recovered in the maximum likelihood phylogeny. The scale bar represents the average number of substitutions per site. For details on analyses, see Materials and Methods. Red crosses indicate complete loss of methanogenesis. Colored spots indicate presence of the genes of the respective methanogenic pathways/reactions as indicated by the colored boxes in figure 1. Crosses within spots indicate that the enzymes are present but not used to perform methanogenesis from H2/CO2 and/or from methylated compounds. Genomes with a green spot harbor the genes needed to use at least one type of methyl compound. (b) Maximum likelihood phylogeny of methanogens based on a concatenation of McrA-B-C-D-G protein sequences comprising 1,159 amino acid positions. Values at nodes represent bootstrap proportions calculated based on 100 resamplings of the original alignment and Bayesian posterior probabilities. For clarity, only the values corresponding to the monophyly of orders and their evolutionary relationships are shown. The scale bar represents the average number of substitutions per site. For details on analyses, see Materials and Methods.