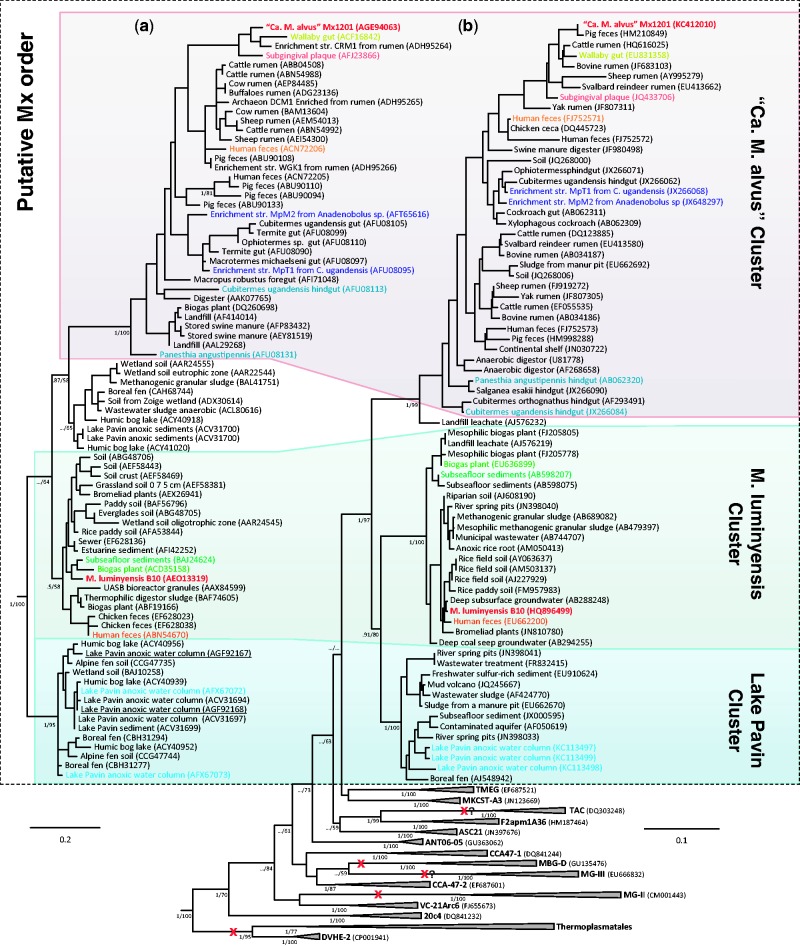
Fig. 3.—
Diversity of the Mx order and its evolutionary relationships with other uncultured lineages. (a) Maximum likelihood McrA phylogeny of the candidate Mx order. (b) Maximum likelihood 16S rRNA phylogeny showing the uncultured lineages branching between the Mx lineage and the Thermoplasmatales/DVHE2, MG-II, and MBG-D lineages. Both trees were rooted by using six representatives of Methanobacteriales and Methanococcales that are not shown for clarity. Trees were calculated from a data set of 138 unambiguously aligned amino acid positions (McrA) and 1,113 unambiguously aligned nucleic acid positions (16S rRNA). Values at nodes represent bootstrap support calculated on 100 resamplings of the original data set and Bayesian posterior probabilities calculated by Bayesian analysis. For clarity, only the values corresponding to the monophyly of lineages and their evolutionary relationships are shown. “../” symbols indicate that the corresponding node was not recovered in the Bayesian phylogeny. The scale bar represents the average number of substitutions per site. For details on data set construction and tree calculation, see Materials and Methods. The dotted box delineates the candidate Mx order. Sequences from the complete genomes of “Candidatus Methanomethylophilus alvus” and Methanomassiliicoccus luminyensis are indicated in red; other colored sequences indicate McrA and 16S rRNA sequences retrieved from the same sample in previous studies. Sequences corresponding to the Lake Pavin sample from which we retrieved Mt coding gene sequences are underlined. Other than the already discussed losses of methanogenesis in the Thermoplasmatales/DVHE2, MG-II lineages, and MBG-D, additional putative losses can be inferred (red crosses with a question mark) in two uncultured lineages based on the environment from which the corresponding sequences were retrieved: oxygenated water column for MG-III; oxygenated and extremely acidic environments (pH <3) for TAC. Lineages 20c4, VC-21Arc6, CCA-47 (divided in two subgroup -1 and -2), ANT06-05, ASC21, F2apm1A36, and MKCST-A3 were derived from the July 2012 version (v.111) of the Silva Database (http://www.arb-silva.de/) and are named according to the clone definition. TMEG, Terrestrial Miscellaneous Euryarchaeotal Group; MBG-D, Marine Benthic Group D; MG-III, Marine Group III; MG-II, Marine Group II; TAC, Thermoplasmatales-related Acidic Cluster.