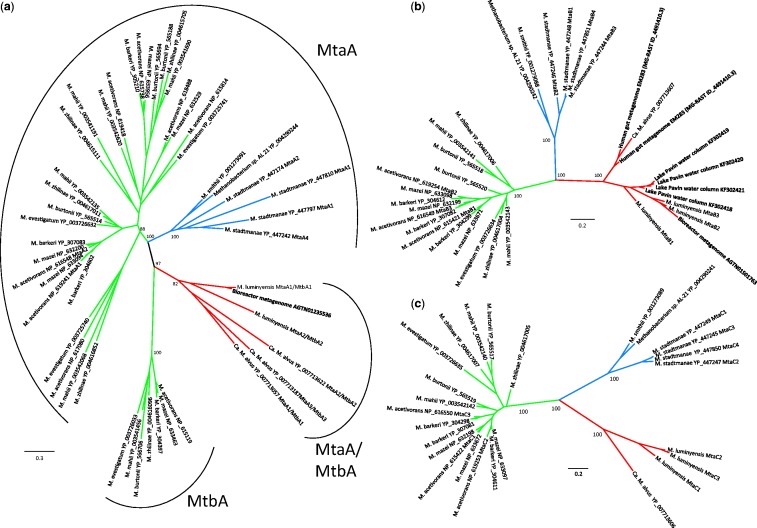
Fig. 5.—
Phylogeny of MtaA/MtbA, MtaB, and MtaC. Unrooted maximum likelihood phylogenies of (a) MtaA/MtbA, (b) MtaB, and (c) MtaC. The Methanosarcinales are indicated in green, the Methanobacteriales in blue, and the Mx lineage in red. Sequences in bold belong to uncultivated microorganisms. Of the environmental sequences from Lake Pavin obtained in this study, only MtaB could be included in phylogenetic analysis because the others were too partial. Also, the MtaC homolog from the bioreactor metagenome was too partial to be included in the tree. However, these are clearly affiliated with the Mx lineage. Values at nodes represent bootstrap support calculated based on 100 resamplings of the original data set. For clarity, only the values corresponding to the monophyly of lineages and their evolutionary relationships are shown. The scale bar represents the average number of substitutions per site. For details on data set construction and tree calculation, see Materials and Methods.