Abstract
Oxidative phosphorylation (OXPHOS), the major energy-producing pathway in aerobic organisms, includes protein subunits encoded by both mitochondrial (mt) and nuclear (nu) genomes. How these independent genomes have coevolved is a long-standing question in evolutionary biology. Although mt genes evolve faster than most nu genes, maintenance of OXPHOS structural stability and functional efficiency may involve correlated evolution of mt and nu OXPHOS genes. The nu OXPHOS genes might be predicted to exhibit accelerated evolutionary rates to accommodate the elevated substitution rates of mt OXPHOS subunits with which they interact. Evolutionary rates of nu OXPHOS genes should, therefore, be higher than that of nu genes that are not involved in OXPHOS (nu non-OXPHOS). We tested the compensatory evolution hypothesis by comparing the evolutionary rates (synonymous substitution rate dS and nonsynonymous substitution rate dN) among 13 mt OXPHOS genes, 60 nu OXPHOS genes, and 77 nu non-OXPHOS genes in vertebrates (7 fish and 40 mammal species). The results from a combined analysis of all OXPHOS subunits fit the predictions of the hypothesis. However, results from two OXPHOS complexes did not fit this pattern when analyzed separately. We found that the dN of nu OXPHOS genes for “core” subunits (those involved in the major catalytic activity) was lower than that of “noncore” subunits, whereas there was no significant difference in dN between genes for nu non-OXPHOS and core subunits. This latter finding suggests that compensatory changes play a minor role in the evolution of OXPHOS genes and that the observed accelerated nu substitution rates are due largely to reduced functional constraint on noncore subunits.
Keywords: oxidative phosphorylation, evolutionary rates, cytonuclear coevolution, nonsynonymous substitution, dN, synonymous substitution, dS
Introduction
The oxidative phosphorylation (OXPHOS) system is unique among fundamental metabolic pathways in animals (aside from maintenance and gene expression of mitochondria) in that functional OXPHOS complexes are composed of subunits encoded by two different genomes. The different hereditary modes of the mitochondrial (mt) and nuclear (nu) genomes substantially increase the complexity of maintaining functional interactions among OXPHOS subunits. The role of OXPHOS in the fundamental process of aerobic ATP production means that slight perturbations to inter-genomic coordination result in a wide range of human pathologies (Wallace 2010). In addition, minimal divergence of OXPHOS subunits may result in reproductive incompatibility among closely related populations (Rand et al. 2004). Consequently, the successful interaction of OXPHOS proteins encoded by mt and nu genomes plays a central role in fundamental processes at the cellular, organismal, and population levels of biological organization. The OXPHOS system provides a rare opportunity to investigate the coevolution of subunits that must successfully interact in electron transport and ATP production, yet are subject to potentially the very different evolutionary forces that govern the mt and nu genomes.
OXPHOS is a complex system composed of the electron transport chain and ATP synthase. It produces about 80% of energy in the form of ATP in almost all eukaryote cells and is the major role-player making mitochondria the “powerhouse of the cell.” Meantime, reactive oxygen species, the by-product of normal metabolism, can damage DNA, cell membranes and lipid, decrease bioenergetic efficiency, and thus lead to age, disease, and death in humans (Ballard and Melvin 2010; Wallace 2010). The OXPHOS apparatus includes five complexes (I–V), each composed of between 4 and 30+ subunits (De Grassi et al. 2005). Complexes I–IV transfer electrons through a series of redox reactions coupled to the transport of protons into the intermembrane space. Complex V employs the resulting electrochemical gradient to phosphorylate ADP to ATP. Like most multimeric protein complexes, successful OXPHOS function requires that subunits maintain structures that allow for specific protein–protein interactions, both within and among complexes. As a result, amino acid residues at many sites are expected to be tightly constrained. An added level of complexity arises from the fact that the composition of four of the five OXPHOS complexes includes subunits encoded by both mt and nu genomes. Any mismatch between mt and nu genomes likely results in a slowing of electron transfer, which increases reactivity of electron with oxygen, corresponding to a rise in free-radical leak (Lane 2011). Evolution of OXPHOS must therefore be coordinated among a large number of interacting subunits and among genomes, and compensatory amino acid substitutions may, in some cases, be favored to maintain function.
The OXPHOS pathway is localized in the mt inner membrane. Complex I (NADH: quinone oxidoreductase) and complex II (succinate dehydrogenase) receive electrons from reducing equivalents (NADH and FADH2) produced by the Krebs cycle. The electrons are transferred to ubiquinone, which freely diffuses within the mt inner membrane and transfers electrons to complex III (cytochrome bc1). Complex III transfers electrons from ubiquinol to cytochrome c. This small protein is localized on the intermembrane space side of the mt inner membrane. It transfers electrons to complex IV (cytochrome c oxidase), where oxygen is reduced to water. Coupled with the process of electron transport, complexes I, III, and IV transport protons to the intermembrane space. The generated proton gradient across the mt inner membrane drives ATP synthesis in complex V (ATP synthase) (Mitchell 1961; Saraste 1999; Boekema and Braun 2007).
The mt genome has a constant gene content composition in all metazoan species (with few exceptions), including 13 protein-coding genes, 2 rRNA genes, and 22 tRNA genes. The rRNA and tRNA genes are involved in mt protein synthesis, whereas the 13 protein-coding genes all encode components of OXPHOS (Shadel and Clayton 1997; Gray et al. 1999). At least 60 OXPHOS subunits are encoded in the nu genome. Despite the fundamental importance of OXPHOS to aerobic life, mt protein-coding genes are not highly conserved and evolve at rates that is 5–50 times that of typical nu genes in vertebrates (Lynch 2007). This increased rate has been observed in a range of animal species, including primates (Brown et al. 1979), Galapagos tortoises (Caccone et al. 2004), lice (Johnson et al. 2003), Drosophila (Haag-Liautard et al. 2008), wasps (Kaltenpoth et al. 2012), and nematodes (Denver et al. 2000). The rapid evolutionary rate of mt genomes may be due to their cell cycle-independent replication of mtDNA (Bogenhagen and Clayton 1977), increased exposure to mutagenic oxygen radical species (Beal 1996), lack of protective histones, and limited DNA repair capacity (Croteau and Bohr 1997). Moreover, the mt genome differs from the nu genome in being effectively haploid, maternally inherited, and exhibiting little recombination (Moritz et al. 1987; Lightowlers et al. 1997; Ballard and Melvin 2010). Haploid maternal inheritance means that the effective population size for mt genes is approximately one-fourth that of nu autosomal genes, increasing the fixation rate of polymorphic sites (Gabriel et al. 1993; Birky 2001). In addition, the lack of recombination in animal mitochondria reduces the ability to purge deleterious mutations (Gabriel et al. 1993). Incompatibility between mt and nu genomes has been shown to reduce hybrid fitness in copepods (Willett and Burton 2001), Drosophila (Sackton et al. 2003), yeast (Zeyl et al. 2005), and wasps (Niehuis et al. 2008), leading to the suggestion that mt–nu incompatibility may be an important contributor to the process of speciation (Gershoni et al. 2009) and the evolution of two separate sexes (Hadjivasiliou et al. 2012).
Among interacting proteins, amino acid substitutions may be deleterious if they affect domains involved in interaction, yet there is the potential for compensatory changes at interacting amino acid sites to reduce or eliminate any negative effects. Such interacting sites are generally proximal to each other in the three-dimensional protein structure (Pazos and Valencia 2008). Under the compensatory model, substitutions that may destabilize protein structures or inhibit function could be compensated by a subsequent (or simultaneous) substitution at an adjacent site (Pollock et al. 1999). In the OXPHOS system, the elevated evolutionary rate of mtDNA suggests that deleterious substitutions occur more frequently in mt genes and compensatory changes may then occur in proximal sites of nu-encoded proteins. In bacteria (Sharp and Li 1987), Drosophila (Comeron and Kreitman 1998; Dunn et al. 2001), and mammals (Wolfe and Sharp 1993), the rate of nonsynonymous substitutions (dN) is positively correlated with the synonymous substitution rate (dS) and both dN and dS are higher in mt than nu genes. If nonsynonymous substitutions of mt genes drive corresponding nonsynonymous substitutions of nu OXPHOS genes, we expect to see the acceleration of dN in nu OXPHOS genes relative to most nu genes not involved in OXPHOS (nu non-OXPHOS).
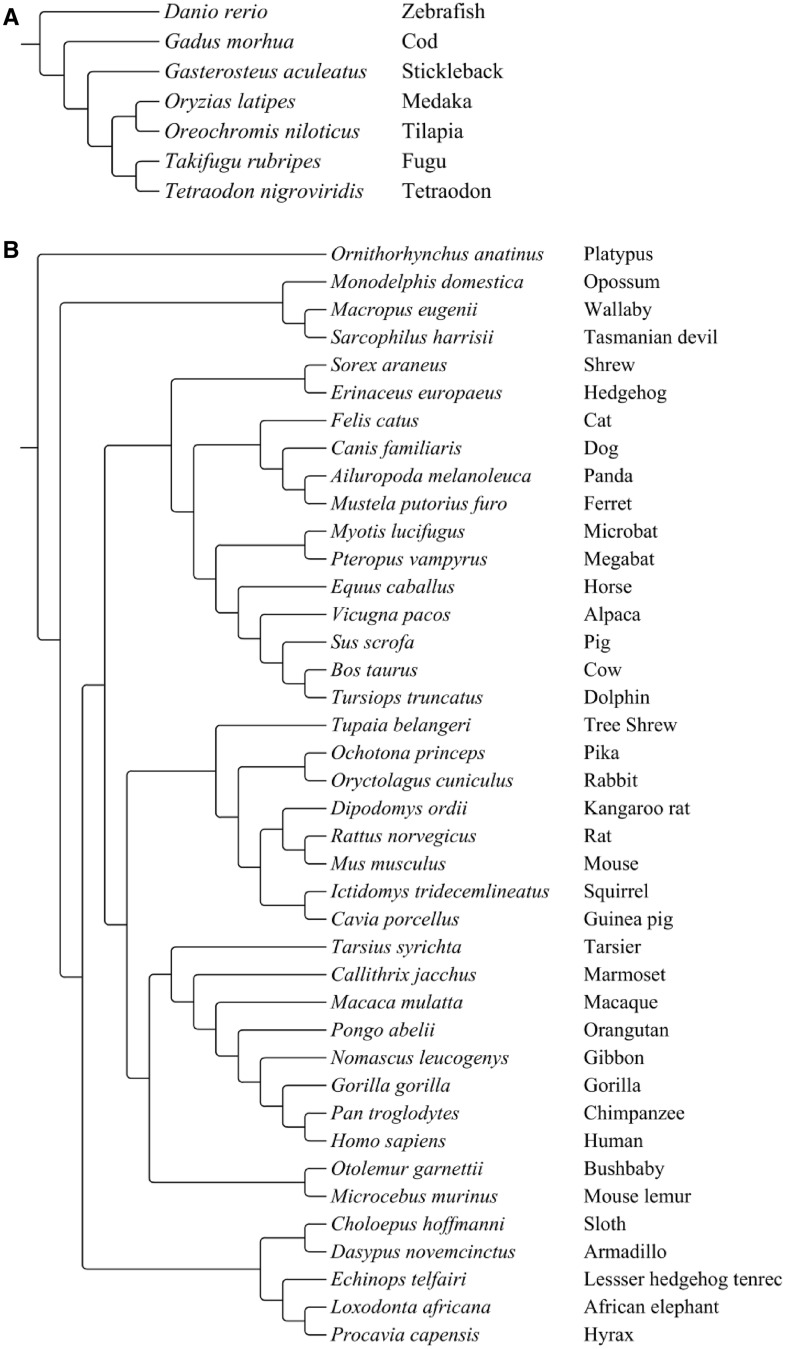
A few recent investigations have addressed mt–nu coevolution at the molecular level but have been restricted to a few genes or a small number of species (e.g., nu genes CYC1 and UQCRFS1, and mt gene MT-CYTB [Willett and Burton 2001], or genes involved in OXPHOS complex IV [Goldberg et al. 2003]). Here, we describe a broader study of 73 OXPHOS genes and a comparison between OXPHOS and 77 non-OXPHOS housekeeping genes. Under a compensatory evolution scenario, we predicted that dN of mt OXPHOS > nu OXPHOS > nu non-OXPHOS genes. We tested the compensatory substitution model by comparing the evolutionary rates (both dS and dN) of 13 mt protein-coding genes (mt OXPHOS genes), 60 nu OXPHOS genes, and 77 non-OXPHOS genes in 47 vertebrate species, including 7 fishes and 40 mammals. The seven fish species are phylogenetically disparate, spanning some 250 Myr of evolutionary history (Betancur-R et al. 2013) (fig. 1A), whereas most of the mammal lineages emerged within the last 80 Myr (Meredith et al. 2011) (fig. 1B).
Fig. 1.—
Phylogeny of the seven fishes (Betancur-R et al. 2013) (A) and 40 mammals (Meredith et al. 2011) (B) used in this study.
Materials and Methods
This study employed genome sequences of 7 teleost fishes and 40 mammals from which we acquired the coding regions of 13 mt OXPHOS and 60 nu OXPHOS genes. For comparison, we sampled 77 housekeeping genes (Warrington et al. 2000; Amsterdam et al. 2004). These were considered appropriate “control” comparators as OXPHOS genes contribute to critical function and should be highly expressed in virtually all cell types. These were chosen randomly with respect to genome location and represent a wide range of functions. They should be broadly representative of functionally important and relatively highly expressed loci in the nu genome. All sequences were downloaded from the Ensembl Genome Browser (www.ensembl.org, last accessed September 11, 2013) using a pipeline procedure (Vilella et al. 2009) (supplementary table S1, Supplementary Material online). A phylogenetic gene tree was used to identify orthologous and paralogous sequences so that we used only the former. Each gene sequence was then realigned using Geneious Pro v.5.6 (Biomatters, Ltd.). A few genes were unavailable in the genome sequences; however, at least 87% of the genes were present in 33 species. The majority of the missing genes were not core OXPHOS subunits whereas the specific genes that were absent varied among species.
Phylogenetic relationships of the fish species followed that of Betancur-R et al. (2013) (fig. 1A), whereas the phylogeny of mammals was from Meredith et al. (2011) (fig. 1B). The program CODEML implemented in the computer software PAML v.4.7 (Yang 2007) was used to estimate dN and dS for each gene. The site-model M0 (model: 0, NSsites: 0, and fix-omega: 0) was used when estimating dN and dS. For each gene, the estimated substitution rates are the sum of dN or dS values from each branch of the tree, so total rate values will vary with the number of taxa in the tree. The intent here was not to compare the absolute values between fishes and mammals, but instead to examine the patterns among mt OXPHOS genes, nu OXPHOS genes, and non-OXPHOS genes within taxonomic groups (where the number of branches was identical). Codon frequencies were estimated from the average nucleotide frequencies at three codon positions. To avoid problems arising from local optima (Yang and Nielsen 1998), replicate runs with three different starting values of ω (0.3, 0.9, and 4.3) were used. Different starting ω yielded similar, and in many cases identical, dS and dN estimates for each gene among three runs (supplementary table S2, Supplementary Material online). The average dS and dN from the three runs was used in the subsequent analysis.
We tested the hypothesis that there are differences in dS and dN among three groups of genes: 13 mt OXPHOS genes, 60 nu OXPHOS genes, and 77 non-OXPHOS genes. A Shapiro–Wilks test was performed to assess the normality of dS and dN for all genes and Bartlett’s test was performed to evaluate homogeneity of variance. Based on these results, a Kruskal–Wallis test followed by post hoc Tukey’s HSD tests were used to assess differences among the groups. A Wilcoxon signed-rank test was performed to evaluate differences between mt and nu OXPHOS genes in each of the five OXPHOS complexes. All statistical analyses were performed in R (R Development Core Team 2008).
Each of the OXPHOS complexes is composed of multiple subunits and each subunit may be more or less important to the core catalytic activity of the complex. We partitioned OXPHOS genes into “core” vs. “noncore,” where core genes encode subunits that retain structural, functional, and sequence homology with prokaryotic systems. For example, unlike human complex I, which consists of 43 subunits, bacterial complex I consists of 14 subunits. These 14 subunits all have homologs in human complex I, and thus are referred as core subunits. These include seven nu OXPHOS genes (NDUFS1–3, NDUFS7–8, and NDUFV1–2) and seven mt OXOPHOS genes (MT-ND1–6, MT-ND4L) (Janssen et al. 2006). Similarly, in complex IV, bovine mt OXPHOS genes (MT-COX1–3) share high similarity to their bacterial counterparts and can be considered the functional core of the eukaryotic oxidase (Richter and Ludwig 2003). Complex II is only composed of four nu subunits (SDHA-D), all of which share high sequence similarity with Escherichia coli (Cecchini 2003). In complex V, six human subunits have bacterial homologs (ATP5A1, ATP5B, ATP5C1, ATP5D, ATP5F1, and ATP5G3) (Yoshida et al. 2001) and were categorized as core subunits. Because E. coli does not have a homolog of complex III (Lenn et al. 2008), none of the nu OXPHOS genes in complex III were categorized as encoding core subunits.
Results
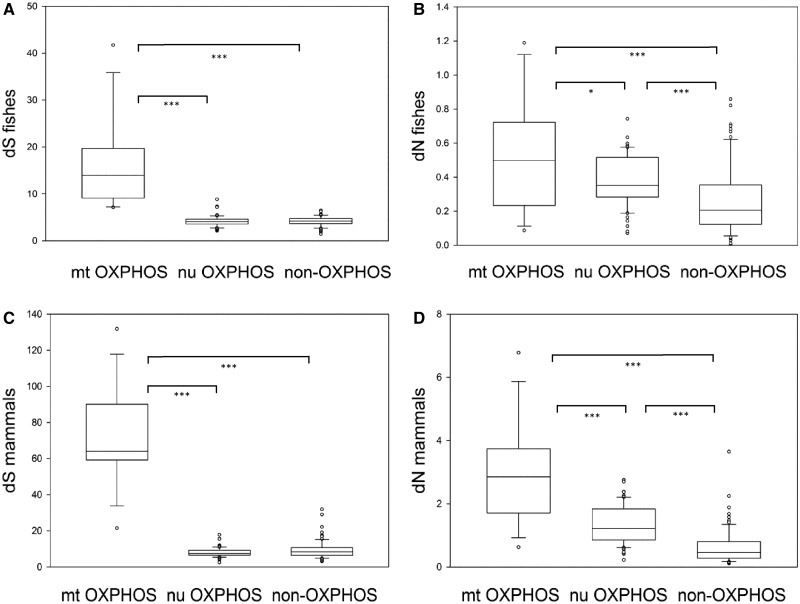
We estimated the total synonymous and nonsynonymous substitution rates on each tree for fishes (fig. 1A) and mammals (fig. 1B) separately using maximum likelihood methods. Because neither dS nor dN was normally distributed and each exhibited significant variance heterogeneity, nonparametric statistics were used to compare the different groups of genes. We found significant differences (Kruskal–Wallis) in dS and dN among the three groups of genes: 13 mt OXPHOS genes, 60 nu OXPHOS genes, and 77 non-OXPHOS genes (table 1). The dS of mt OXPHOS genes was significantly higher than that of nu OXPHOS and non-OXPHOS genes in both (fig. 2A and C). The dS of mt and nu genes were 16.02 ± 9.64 and 2.02 ± 0.87, respectively, in fishes and 72.27 ± 26.56 and 8.85 ± 4.15, respectively, in mammals. No significant difference of the dS was detected between nu OXPHOS genes and non-OXPHOS genes (fig. 2A and C). We note that a small number of mt genes had exceptionally high dS values, which may indicate substitutional saturation, leading to violation of the assumptions of the maximum likelihood estimation method. As predicted, the dN of mt OXPHOS genes was significantly higher than that of nu OXPHOS genes, which in turn was significantly higher than that of non-OXPHOS genes (fig. 2B and D).
Table 1.
Statistical Summary for the Comparison of Synonymous Substitution Rate (dS) and Nonsynonymous Substitution Rate (dN) among Mitochondrial Oxidative Phosphorylation (mt OXPHOS), Nuclear OXPHOS (nu OXPHOS), and Non-OXPHOS Genes in 7 Fishes and 40 Mammals
| ML Estimates | Taxa | Comparison | P Value |
|---|---|---|---|
| dS | Fishes | mt OXPHOS > nu OXPHOS | <0.0001 |
| dS | Mammals | mt OXPHOS > nu OXPHOS | <0.0001 |
| dS | Fishes | mt OXPHOS > non-OXPHOS | <0.0001 |
| dS | Mammals | mt OXPHOS > non-OXPHOS | <0.0001 |
| dS | Fishes | nu OXPHOS > non-OXPHOS | 0.987 |
| dS | Mammals | nu OXPHOS > non-OXPHOS | 0.670 |
| dN | Fishes | mt OXPHOS > nu OXPHOS | 0.037 |
| dN | Mammals | mt OXPHOS > nu OXPHOS | <0.0001 |
| dN | Fishes | mt OXPHOS > non-OXPHOS | <0.0001 |
| dN | Mammals | mt OXPHOS > non-OXPHOS | <0.0001 |
| dN | Fishes | nu OXPHOS > non-OXPHOS | 0.0007 |
| dN | Mammals | nu OXPHOS > non-OXPHOS | <0.0001 |
Fig. 2.—
Comparison of synonymous substitution rate (dS) and nonsynonymous substitution rate (dN) among mitochondrial oxidative phosphorylation (mt OXPHOS), nuclear OXPHOS (nu OXPHOS), and non-OXPHOS genes in 7 fishes (A, B) and 40 mammals (C, D). Whisker-ends are at the 5th and 95th percentiles. *P < 0.05, **P < 0.01, ***P < 0.001.
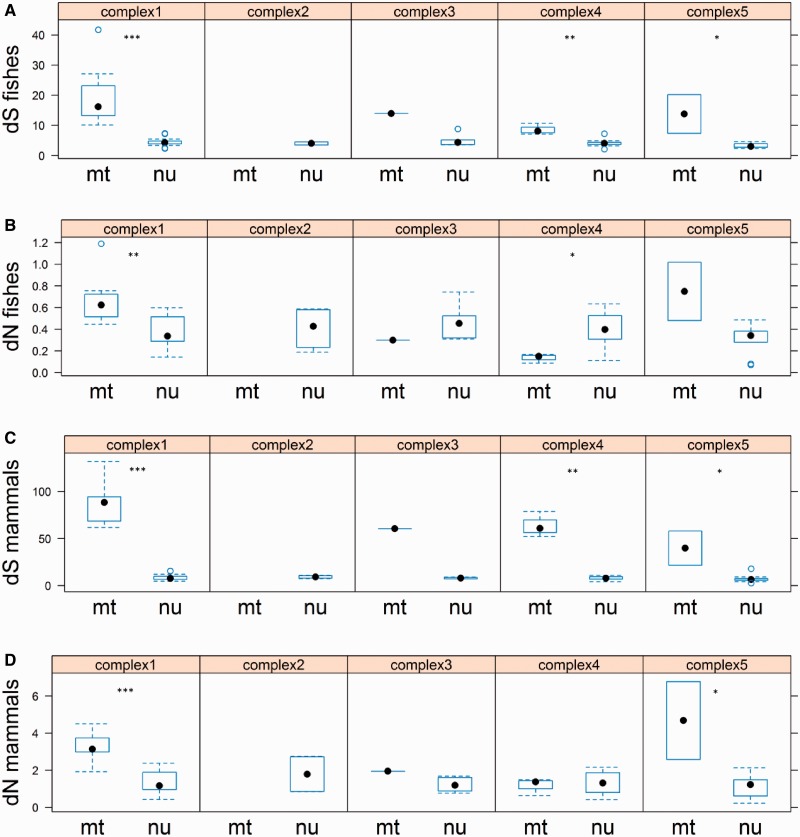
Estimates of dS and dN were assessed for each OXPHOS complexes. The dS of mt genes was higher than that of nu genes in all four complexes (complex II is composed of subunits only encoded by nu genes) (fig. 3A and C). The dN of mt genes was not consistently higher than that of nu genes in all complexes. For example, the dN of mt genes in complex III was lower than that of nu genes in fishes (fig. 3B) whereas higher than that of nu genes in mammals (fig. 3D); the dN of mt genes in complex IV was significantly lower than that of nu genes in fishes (fig. 3B) and lower than that of nu genes in mammals (fig. 3D).
Fig. 3.—
Comparison of synonymous substitution rate (dS) and nonsynonymous substitution rate (dN) between mitochondrial (mt) and nuclear (nu) genes in each complex in 7 fishes (A, B) and 40 mammals (C, D). Complex II is composed of subunits encoded only by nu genes. Whisker-ends are at the 5th and 95th percentiles. *P < 0.05, **P < 0.01, ***P < 0.001.
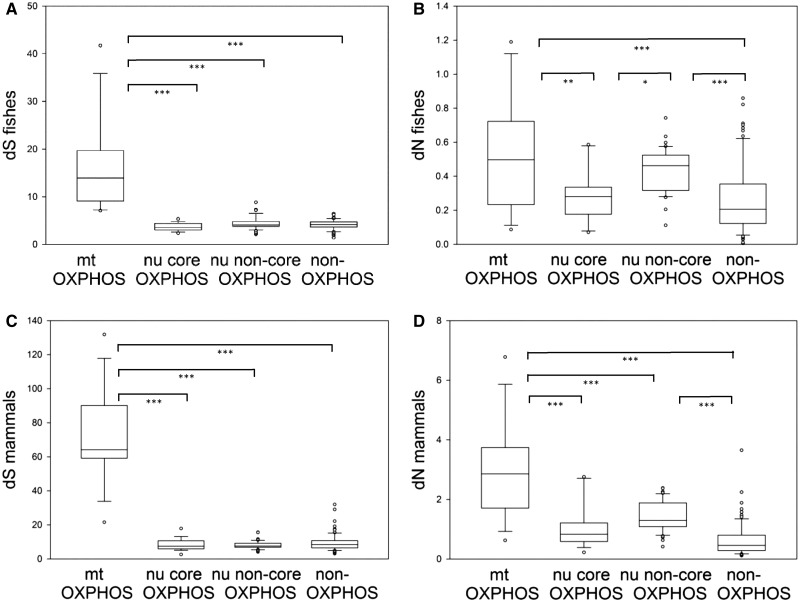
Because of their different functions and interactions, OXPHOS genes may be under different levels of functional constraint, so we separated them into core and noncore groups. Core subunits were defined as those that have bacterial homologs and that participate directly in electron transport or ATP synthesis. After partitioning, dS of mt OXPHOS genes was found to be significantly higher than that of all nu gene groups (fig. 4A and C), whereas the dN of noncore OXPHOS genes was higher than that of core OXPHOS genes (fig. 4B and D). There were no significant differences in dN between nu non-OXPHOS genes and nu core OXPHOS genes (fig. 4B and D, table 2).
Fig. 4.—
Comparison of synonymous substitution rate (dS) and nonsynonymous substitution rate (dN) among mitochondrial oxidative phosphorylation (mt OXPHOS), nuclear core OXPHOS (nu core OXPHOS), nuclear noncore OXPHOS (nu noncore OXPHOS), and non-OXPHOS genes in 7 fishes (A, B) and 40 mammals (C, D). Whisker-ends are at the 5th and 95th percentiles. *P < 0.05, **P < 0.01, ***P < 0.001.
Table 2.
Statistical Summary for the Comparison of Synonymous Substitution Rate (dS) and Nonsynonymous Substitution Rate (dN) among Mitochondrial Oxidative Phosphorylation (mt OXPHOS), Nuclear Core OXPHOS (nu core OXPHOS), Nuclear Noncore OXPHOS (nu noncore OXPHOS), and Non-OXPHOS Genes in 7 Fishes and 40 Mammals
| ML Estimates | Taxa | Comparison | P Value |
|---|---|---|---|
| dS | Fishes | mt OXPHOS > nu core OXPHOS | <0.0001 |
| dS | Mammals | mt OXPHOS > nu core OXPHOS | <0.0001 |
| dS | Fishes | mt OXPHOS > nu noncore OXPHOS | <0.0001 |
| dS | Mammals | mt OXPHOS > nu noncore OXPHOS | <0.0001 |
| dS | Fishes | mt OXPHOS > non-OXPHOS | <0.0001 |
| dS | Mammals | mt OXPHOS > non-OXPHOS | <0.0001 |
| dS | Fishes | nu core OXPHOS < nu noncore OXPHOS | 0.881 |
| dS | Mammals | nu core OXPHOS > nu noncore OXPHOS | 0.9995 |
| dS | Fishes | nu core OXPHOS < non-OXPHOS | 0.966 |
| dS | Mammals | nu core OXPHOS < non-OXPHOS | 0.967 |
| dS | Fishes | nu noncore OXPHOS > non-OXPHOS | 0.969 |
| dS | Mammals | nu noncore OXPHOS < non-OXPHOS | 0.847 |
| dN | Fishes | mt OXPHOS > nu core OXPHOS | 0.002 |
| dN | Mammals | mt OXPHOS > nu core OXPHOS | <0.0001 |
| dN | Fishes | mt OXPHOS > nu noncore OXPHOS | <0.304 |
| dN | Mammals | mt OXPHOS > nu noncore OXPHOS | <0.0001 |
| dN | Fishes | mt OXPHOS > non-OXPHOS | <0.0001 |
| dN | Mammals | mt OXPHOS > non-OXPHOS | <0.0001 |
| dN | Fishes | nu core OXPHOS < nu noncore OXPHOS | 0.031 |
| dN | Mammals | nu core OXPHOS < nu noncore OXPHOS | 0.193 |
| dN | Fishes | nu core OXPHOS > non-OXPHOS | 0.988 |
| dN | Mammals | nu core OXPHOS > non-OXPHOS | 0.198 |
| dN | Fishes | nu noncore OXPHOS > non-OXPHOS | <0.0001 |
| dN | Mammals | nu noncore OXPHOS > non-OXPHOS | <0.0001 |
Discussion
Given that mt genome is derived from once free-living bacteria, it is clear that most of the bacterial genes have now been removed to the cell nucleus. However, what has not been well resolved is why not all genes have been removed to the nucleus (Alberts et al. 1994). The remained genes in mitochondria thus are the resources for any potential problems involved in the interaction between mt–nu genomes. To infer factors influencing the evolution of mt–nu interactions, we used estimated rates of synonymous and nonsynonymous substitutions among all OXPHOS genes and a broad genomic sample of housekeeping genes. Specifically, we examined the hypothesis of compensatory evolution between mt and nu OXPHOS genes by comparing their evolutionary rates (both dS and dN) relative to non-OXPHOS genes in two distinct vertebrate lineages. We found that the mean dS of mt genes was about 7–9 times higher than that of nu genes. This magnitude is consistent with the well-established pattern of high dS in animal mt genes (Brown et al. 1979; Johnson et al. 2003; Caccone et al. 2004; Lynch 2007; Kaltenpoth et al. 2012). Although the products of nu OXHPOS genes are transported into mitochondria where they function, they are expected to show mean dS similar to the non-OXPHOS genes because the coding sequences reside in the nu genome. Our results show this to be the case and suggest that differences in nonsynonymous rates are due to selective factors rather than different mutation rates.
mt–nu coevolution could generate predictions at different levels. One prediction at the cellular level is that in experimental “transplants” interacting partners should result in diminished functional performance, and this disruption should increase as the level of evolutionary divergence increases (Rand et al. 2004). In mouse (Mus musculus domesticus) cell lines carrying mitochondria from six different murid species spanning 2–12 Myr of divergence, a near-linear association between disruption of respiratory chain function and evolutionary distance has been observed (McKenzie et al. 2003). This phenomenon has also been observed in primates (Kenyon and Moraes 1997), copepods (Willett and Burton 2001), Drosophila (Sackton et al. 2003), and wasps (Niehuis et al. 2008). However, more limited divergence may not lead to disruption of performance. Introgression experiments of Drosophila simulans siII mtDNA type into the sympatric population siIII nu background did not show a difference in catalytic properties of mitochondria, indicating that some naturally occurring mutations in mtDNA can be accommodated by different nu background and mt–nu interaction (Pichaud et al. 2012).
At the DNA sequence level, higher evolutionary rates of mt genes could drive accelerated evolutionary rates of nu genes as compensatory response in the nu genes contributes to the maintenance of function. Several studies of the primate complex IV showed that seven nu OXPHOS genes: COX4I1 (Wu et al. 1997; Wildman et al. 2002), COX7A1H (Schmidt et al. 1999), COX5A (Doan et al. 2004), COX8AL (Osada et al. 2002; Goldberg et al. 2003), COX6B1, COX6C, and COX7C (Doan et al. 2004), together with two mt OXPHOS genes: MT-COX1 (Andrews and Easteal 2000; Wu et al. 2000) and MT-COX2 (Adkins and Honeycutt 1994), have shown accelerated dN in the lineage leading to hominids relative to other primates. However, in these cases it is not clear whether accelerated dN is due to compensatory changes driven by the higher mt rate, or is due to selection for increased OXPHOS efficiency associated with increased brain size in the hominid lineage. This study showed that when all complexes were analyzed together, the dN of 13 mt OXPHOS genes was significantly higher than that of 60 nu OXPHOS genes, and both of these were significantly higher than dN of 77 non-OXPHOS genes. The apparent acceleration dN of nu OXPHOS genes is consistent with a compensatory response to the higher dN of mt OXPHOS genes.
However, this explanation seems tenuous when considering rates in each complex separately. Contrary to expectations, our results showed that the dN of mt genes was not always higher than that of nu genes in each complex. mt genes with higher dN than nu genes were only observed in complexes I and V. In fishes, the dN of mt genes was lower than that of nu genes in complexes III and IV. In mammals, the dN of mt genes was lower than that of nu genes in complex IV, but higher in complex III. Goldberg et al. (2003) found that in the anthropoid lineage, the fast-evolving region (12 N-terminal amino acid residues) of COX8L encodes amino acid sites contacted with MT-COX1 sites in three-dimensional structure. This suggested structurally mediated cytonuclear coevolution, which was driven by faster evolving mt OXPHOS genes. However, we found no evidence of such a pattern in fishes and mammals. Similarly, the only mt gene in complex III (MT-CYTB) showed lower dN than its corresponding nu OXPHOS genes in fishes. Although dN of MT-CYTB was higher than nu genes in complex III in mammals, the difference is not statistically different. These results are clearly counter to expectations under a compensatory model.
One explanation for the lower dN of mt OXPHOS genes in complexes III and IV could be functional constraints on core subunits. MT-CYTB is the only mt gene in complex III and its structure has been shown to be conserved among vertebrates (Kocher et al. 1989). Similarly, MT-COX1 and MT-COX2 have been recognized as the core enzyme catalytic subunits and are conserved from bacteria to bovid (Tsukihara et al. 1996). It therefore seems likely that functional constraints are stronger on core subunits, regardless of which genome encodes them. The higher dN of nu OXPHOS genes relative to non-OXPHOS genes in general is due mainly to higher rates of noncore proteins alone. The more recently derived noncore subunits (or “accessory OXPHOS families”) appear to be important contributors to OXPHOS assembly and/or stabilization (Ugalde et al. 2004), but do not contribute directly to catalytic activity of electron transport and ATP synthesis (De Grassi et al. 2005). Thus, maintenance of primary amino acid sequences of the noncore subunits appears to be less important to OXPHOS function than that of the core subunits and the intensity of selection on these proteins is therefore reduced.
It is possible that different domains within a protein may be under different selective regimes and such domains may have different evolutionary rates. Such differences could be obscured by combining domains and estimating dN for each gene as a whole. For example, a gene with a small but highly functionally constrained domain combined with large relatively unconserved domains would exhibit high dN despite having an important conserved function. Had we obtained high relative dN for core OXPHOS genes, such a result would have been consistent with a compensatory evolution hypothesis but could have been due to such overestimation of dN. However, a salient result here is the relatively low estimates of dN for several mt and core nu OXPHOS genes. It seems unlikely that differential selection on domains could lead to underestimation of dN if the compensatory evolution hypothesis (which predicts elevated dN values) was a major factor influence in OXPHOS evolution.
We cannot exclude the possibility of a few compensatory changes in nu OXPHOS genes as they “keep pace” with changes in mt OXPHOS genes because assessment of evolutionary rates does not allow examination of individual sites. Neither can we reject compensatory changes in mt genes in response to changes in the nu genome; indeed, the rapid rate of mt genome evolution could allow these genes to more readily respond to changes in nu components. Nonetheless, compensatory changes in either direction do not appear to occur with a frequency sufficient to contribute to differences in evolutionary rates among genes or groups of genes. Therefore, despite the necessary interaction of the products of two different genomes, OXPHOS evolution appears to be driven largely by conventional natural selection for functional efficiency acting on individual subunits, regardless of their genome of origin.
Supplementary Material
Supplementary tables S1 and S2 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors thank J.C. Cureton II for helpful discussions and L.J. Weider, J.P. Masly, C.M. Lewis, and R. Knapp for suggestions on manuscript. This work was supported by National Science Foundation award DEB-0732988 (to R.E.B.).
Literature Cited
- Adkins RM, Honeycutt RL. Evolution of the primate cytochrome c oxidase subunit II gene. J Mol Evol. 1994;38:215–231. doi: 10.1007/BF00176084. [DOI] [PubMed] [Google Scholar]
- Alberts B, et al. The molecular biology of the cell. New York, London: Garland Publishing; 1994. [Google Scholar]
- Amsterdam A, et al. Identification of 315 genes essential for early zebrafish development. Proc Natl Acad Sci U S A. 2004;101:12792–12797. doi: 10.1073/pnas.0403929101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews TD, Easteal S. Evolutionary rate acceleration of cytochrome c oxidase subunit I in simian primates. J Mol Evol. 2000;50:562–568. doi: 10.1007/s002390010059. [DOI] [PubMed] [Google Scholar]
- Ballard J, Melvin R. Linking the mitochondrial genotype to the organismal phenotype. Mol Ecol. 2010;19:1523–1539. doi: 10.1111/j.1365-294X.2010.04594.x. [DOI] [PubMed] [Google Scholar]
- Beal M. Mitochondria, free radicals, and neurodegeneration. Curr Opin Neurobiol. 1996;6:661–666. doi: 10.1016/s0959-4388(96)80100-0. [DOI] [PubMed] [Google Scholar]
- Betancur-R R, et al. The tree of life and a new classification of bony fishes. PLoS Curr. 2013 doi: 10.1371/currents.tol.53ba26640df0ccaee75bb165c8c26288. Advance Access published April 18, 2013, doi: 10.1371/currents.tol.53ba26640df0ccaee75bb165c8c26288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birky C. The inheritance of genes in mitochondria and chloroplasts: laws, mechanisms, and models. Annu Rev Genet. 2001;35:125–148. doi: 10.1146/annurev.genet.35.102401.090231. [DOI] [PubMed] [Google Scholar]
- Boekema E, Braun H-P. Supramolecular structure of the mitochondrial oxidative phosphorylation system. J Biol Chem. 2007;282:1–4. doi: 10.1074/jbc.R600031200. [DOI] [PubMed] [Google Scholar]
- Bogenhagen D, Clayton DA. Mouse L cell mitochondrial DNA molecules are selected randomly for replication throughout the cell cycle. Cell. 1977;11:719–727. doi: 10.1016/0092-8674(77)90286-0. [DOI] [PubMed] [Google Scholar]
- Brown W, George M, Wilson A. Rapid evolution of animal mitochondrial DNA. Proc Natl Acad Sci U S A. 1979;76:1967–1971. doi: 10.1073/pnas.76.4.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caccone A, et al. Extreme difference in rate of mitochondrial and nuclear DNA evolution in a large ectotherm, Galápagos tortoises. Mol Phylogenet Evol. 2004;31:794–798. doi: 10.1016/j.ympev.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Cecchini G. Function and structure of complex II of the respiratory chain. Annu Rev Biochem. 2003;72:77–109. doi: 10.1146/annurev.biochem.72.121801.161700. [DOI] [PubMed] [Google Scholar]
- Comeron J, Kreitman M. The correlation between synonymous and nonsynonymous substitutions in Drosophila: mutation, selection or relaxed constraints? Genetics. 1998;150:767–775. doi: 10.1093/genetics/150.2.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croteau D, Bohr V. Repair of oxidative damage to nuclear and mitochondrial DNA in mammalian cells. J Biol Chem. 1997;272:25409–25412. doi: 10.1074/jbc.272.41.25409. [DOI] [PubMed] [Google Scholar]
- De Grassi A, et al. Evolution of nuclearly encoded mitochondrial genes in Metazoa. Gene. 2005;354:181–188. doi: 10.1016/j.gene.2005.03.046. [DOI] [PubMed] [Google Scholar]
- Denver DR, Morris K, Lynch M, Vassilieva LL, Thomas WK. High direct estimate of the mutation rate in the mitochondrial genome of Caenorhabditis elegans. Science. 2000;289:2342–2344. doi: 10.1126/science.289.5488.2342. [DOI] [PubMed] [Google Scholar]
- Doan JW, et al. Coadaptive evolution in cytochrome c oxidase: 9 of 13 subunits show accelerated rates of nonsynonymous substitution in anthropoid primates. Mol Phylogenet Evol. 2004;33:944. doi: 10.1016/j.ympev.2004.07.016. [DOI] [PubMed] [Google Scholar]
- Dunn K, Bielawski J, Yang Z. Substitution rates in Drosophila nuclear genes: implications for translational selection. Genetics. 2001;157:295–305. doi: 10.1093/genetics/157.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel W, Lynch M, Burger R. Muller’s ratchet and mutational meltdowns. Evolution. 1993;47:1744–1757. doi: 10.1111/j.1558-5646.1993.tb01266.x. [DOI] [PubMed] [Google Scholar]
- Gershoni M, Templeton A, Mishmar D. Mitochondrial bioenergetics as a major motive force of speciation. Bioessays. 2009;31:642–650. doi: 10.1002/bies.200800139. [DOI] [PubMed] [Google Scholar]
- Goldberg A, et al. Adaptive evolution of cytochrome c oxidase subunit VIII in anthropoid primates. Proc Natl Acad Sci U S A. 2003;100:5873–5878. doi: 10.1073/pnas.0931463100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MW, Burger G, Lang BF. Mitochondrial evolution. Science. 1999;283:1476–1481. doi: 10.1126/science.283.5407.1476. [DOI] [PubMed] [Google Scholar]
- Haag-Liautard C, et al. Direct estimation of the mitochondrial DNA mutation rate in Drosophila melanogaster. PLoS Biol. 2008;6:e204. doi: 10.1371/journal.pbio.0060204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjivasiliou Z, Pomiankowski A, Seymour RM, Lane N. Selection for mitonuclear co-adaptation could favour the evolution of two sexes. Proc R Soc Lond B Biol Sci. 2012;279:1865–1872. doi: 10.1098/rspb.2011.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen RJ, Nijtmans LG, van den Heuvel LP, Smeitink JA. Mitochondrial complex I: structure, function and pathology. J Inherit Metab Dis. 2006;29:499–515. doi: 10.1007/s10545-006-0362-4. [DOI] [PubMed] [Google Scholar]
- Johnson K, et al. Dramatically elevated rate of mitochondrial substitution in lice (Insecta: Phthiraptera) Mol Phylogenet Evol. 2003;26:231–242. doi: 10.1016/s1055-7903(02)00342-1. [DOI] [PubMed] [Google Scholar]
- Kaltenpoth M, et al. Accelerated evolution of mitochondrial but not nuclear genomes of Hymenoptera: new evidence from crabronid wasps. PLoS One. 2012;7:e32826. doi: 10.1371/journal.pone.0032826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon L, Moraes C. Expanding the functional human mitochondrial DNA database by the establishment of primate xenomitochondrial cybrids. Proc Natl Acad Sci U S A. 1997;94:9131–9135. doi: 10.1073/pnas.94.17.9131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocher TD, et al. Dynamics of mitochondrial DNA evolution in animals: amplification and sequencing with conserved primers. Proc Natl Acad Sci U S A. 1989;86:6196–6200. doi: 10.1073/pnas.86.16.6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane N. Mitonuclear match: optimizing fitness and fertility over generations drives ageing within generations. Bioessays. 2011;33(11):860–869. doi: 10.1002/bies.201100051. [DOI] [PubMed] [Google Scholar]
- Lenn T, Leake M, Mullineaux C. Are Escherichia coli OXPHOS complexes concentrated in specialized zones within the plasma membrane? Biochem Soc Trans. 2008;36:1032–1036. doi: 10.1042/BST0361032. [DOI] [PubMed] [Google Scholar]
- Lightowlers R, Chinnery P, Turnbull D, Howell N. Mammalian mitochondrial genetics: heredity, heteroplasmy and disease. Trends Genet. 1997;13:450–455. doi: 10.1016/s0168-9525(97)01266-3. [DOI] [PubMed] [Google Scholar]
- Lynch M. The origins of genome architecture. 2007 Sinauer Associates Inc. [Google Scholar]
- McKenzie M, Chiotis M, Pinkert CA, Trounce IA. Functional respiratory chain analyses in murid xenomitochondrial cybrids expose coevolutionary constraints of cytochrome b and nuclear subunits of complex III. Mol Biol Evol. 2003;20:1117–1124. doi: 10.1093/molbev/msg132. [DOI] [PubMed] [Google Scholar]
- Meredith RW, et al. Impacts of the cretaceous terrestrial revolution and KPg extinction on mammal diversification. Science. 2011;334:521–524. doi: 10.1126/science.1211028. [DOI] [PubMed] [Google Scholar]
- Mitchell P. Coupling of phosphorylation to electron and hydrogen transfer by a chemi-osmotic type of mechanism. Nature. 1961;191:144–148. doi: 10.1038/191144a0. [DOI] [PubMed] [Google Scholar]
- Moritz C, Dowling T, Brown W. Evolution of animal mitochondrial DNA: relevance for population biology and systematics. Annu Rev Ecol Syst. 1987;18:269–292. [Google Scholar]
- Niehuis O, Judson A, Gadau J. Cytonuclear genic incompatibilities cause increased mortality in male F2 hybrids of Nasonia giraulti and N. vitripennis. Genetics. 2008;178:413–426. doi: 10.1534/genetics.107.080523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osada N, et al. Search for genes positively selected during primate evolution by 5′-end-sequence screening of cynomolgus monkey cDNAs. Genomics. 2002;79:657–662. doi: 10.1006/geno.2002.6753. [DOI] [PubMed] [Google Scholar]
- Pazos F, Valencia A. Protein co-evolution, co-adaptation and interactions. EMBO J. 2008;27:2648–2655. doi: 10.1038/emboj.2008.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichaud N, Ballard JWO, Tanguay RM, Blier PU. Naturally occurring mitochondrial DNA haplotypes exhibit metabolic differences: insight into functional properties of mitochondria. Evolution. 2012;66:3189–3197. doi: 10.1111/j.1558-5646.2012.01683.x. [DOI] [PubMed] [Google Scholar]
- Pollock D, Taylor W, Goldman N. Coevolving protein residues: maximum likelihood identification and relationship to structure. J Mol Biol. 1999;287:187–198. doi: 10.1006/jmbi.1998.2601. [DOI] [PubMed] [Google Scholar]
- Rand D, Haney R, Fry A. Cytonuclear coevolution: the genomics of cooperation. Trends Ecol Evol. 2004;19:645–653. doi: 10.1016/j.tree.2004.10.003. [DOI] [PubMed] [Google Scholar]
- R Development Core Team. R: a language and environment for statistical computing. Vienna (Austria): R Foundation for Statistical Computing; 2013. [cited 2013 September 11]. Available from: http://www.R-project.org. [Google Scholar]
- Richter O-M, Ludwig B. Cytochrome c oxidase—structure, function, and physiology of a redox-driven molecular machine. Rev Physiol Biochem Pharmacol. 2003;147:47–74. doi: 10.1007/s10254-003-0006-0. [DOI] [PubMed] [Google Scholar]
- Sackton T, Haney R, Rand D. Cytonuclear coadaptation in Drosophila: disruption of cytochrome c oxidase activity in backcross genotypes. Evolution. 2003;57:2315–2325. doi: 10.1111/j.0014-3820.2003.tb00243.x. [DOI] [PubMed] [Google Scholar]
- Saraste M. Oxidative phosphorylation at the fin de siècle. Science. 1999;283:1488–1493. doi: 10.1126/science.283.5407.1488. [DOI] [PubMed] [Google Scholar]
- Schmidt TR, Goodman M, Grossman LI. Molecular evolution of the COX7A gene family in primates. Mol Biol Evol. 1999;16:619–626. doi: 10.1093/oxfordjournals.molbev.a026144. [DOI] [PubMed] [Google Scholar]
- Shadel G, Clayton D. Mitochondrial DNA maintenance in vertebrates. Annu Rev Biochem. 1997;66:409–435. doi: 10.1146/annurev.biochem.66.1.409. [DOI] [PubMed] [Google Scholar]
- Sharp P, Li W. The rate of synonymous substitution in enterobacterial genes is inversely related to codon usage bias. Mol Biol Evol. 1987;4:222–230. doi: 10.1093/oxfordjournals.molbev.a040443. [DOI] [PubMed] [Google Scholar]
- Tsukihara T, et al. The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 A. Science. 1996;272:1136–1144. doi: 10.1126/science.272.5265.1136. [DOI] [PubMed] [Google Scholar]
- Ugalde C, Janssen RJ, van den Heuvel LP, Smeitink JA, Nijtmans LG. Differences in assembly or stability of complex I and other mitochondrial OXPHOS complexes in inherited complex I deficiency. Hum Mol Genet. 2004;13:659–667. doi: 10.1093/hmg/ddh071. [DOI] [PubMed] [Google Scholar]
- Vilella AJ, et al. EnsemblCompara GeneTrees: complete, duplication-aware phylogenetic trees in vertebrates. Genome Res. 2009;19:327–335. doi: 10.1101/gr.073585.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC. Mitochondrial DNA mutations in disease and aging. Environ Mol Mutagen. 2010;51:440–450. doi: 10.1002/em.20586. [DOI] [PubMed] [Google Scholar]
- Warrington JA, Nair A, Mahadevappa M, Tsyganskaya M. Comparison of human adult and fetal expression and identification of 535 housekeeping/maintenance genes. Physiol Genomics. 2000;2:143–147. doi: 10.1152/physiolgenomics.2000.2.3.143. [DOI] [PubMed] [Google Scholar]
- Wildman DE, Wu W, Goodman M, Grossman LI. Episodic positive selection in ape cytochrome c oxidase subunit IV. Mol Biol Evol. 2002;19:1812–1815. doi: 10.1093/oxfordjournals.molbev.a004005. [DOI] [PubMed] [Google Scholar]
- Willett C, Burton R. Viability of cytochrome c genotypes depends on cytoplasmic backgrounds in Tigriopus californicus. Evolution. 2001;55:1592–1599. doi: 10.1111/j.0014-3820.2001.tb00678.x. [DOI] [PubMed] [Google Scholar]
- Wolfe K, Sharp P. Mammalian gene evolution: nucleotide sequence divergence between mouse and rat. J Mol Evol. 1993;37:441–456. doi: 10.1007/BF00178874. [DOI] [PubMed] [Google Scholar]
- Wu W, Goodman M, Lomax MI, Grossman LI. Molecular evolution of cytochrome c oxidase subunit IV: evidence for positive selection in simian primates. J Mol Evol. 1997;44:477–491. doi: 10.1007/pl00006172. [DOI] [PubMed] [Google Scholar]
- Wu W, Schmidt TR, Goodman M, Grossman LI. Molecular evolution of cytochrome c oxidase subunit I in primates: is there co-evolution between mitochondrial and nuclear genomes? Mol Phylogenet Evol. 2000;17:294–304. doi: 10.1006/mpev.2000.0833. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R. Synonymous and nonsynonymous rate variation in nuclear genes of mammals. J Mol Evol. 1998;46:409–418. doi: 10.1007/pl00006320. [DOI] [PubMed] [Google Scholar]
- Yoshida M, Muneyuki E, Hisabori T. ATP synthase—a marvellous rotary engine of the cell. Nat Rev Mol Cell Biol. 2001;2:669–677. doi: 10.1038/35089509. [DOI] [PubMed] [Google Scholar]
- Zeyl C, Andreson B, Weninck E. Nuclear-mitochondrial epistasis for fitness in Saccharomyces cerevisiae. Evolution. 2005;59:910–914. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.