Table 2.
Key Statistics for Simulated Data Sets
| % Duplications |
|||||
|---|---|---|---|---|---|
| 0 | 10 | 20 | 30 | 40 | |
| Parameters values | |||||
| No. of sequences | 1,000 | ||||
| Distr. of seq. length | 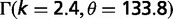 |
||||
| Min. sequence length | 50 | ||||
| Substitution model | WAG | ||||
| Insertion and deletion rate | 0.000125 | ||||
| Gene duplication rate | 0 | 0.003 | 0.0056 | 0.009 | 0.0125 |
| Gene loss rate | 0 | 0.003 | |||
| No. of species | 30 | ||||
| Key statistics | |||||
| Seq. length (mean) | 316.6 | 326.4 | 323.3 | 325.0 | 320.3 |
| Seq. length (stderr) | 201.7 | 211.6 | 207.4 | 213.1 | 203.6 |
| Avg. % gap chars in MSA | 24.27 | 23.25 | 24.64 | 26.23 | 28.65 |
| Variance of % gap chars | 58.0 | 62.8 | 66.4 | 72.4 | 80.5 |
| Total species tree length | 763.6 | ||||
| Minimum species tree height | 31.70 | ||||
| Maximum species tree height | 77.80 | ||||
| Average species tree height | 41.36 | ||||
| Average distance between species pairs | 72.60 | ||||
