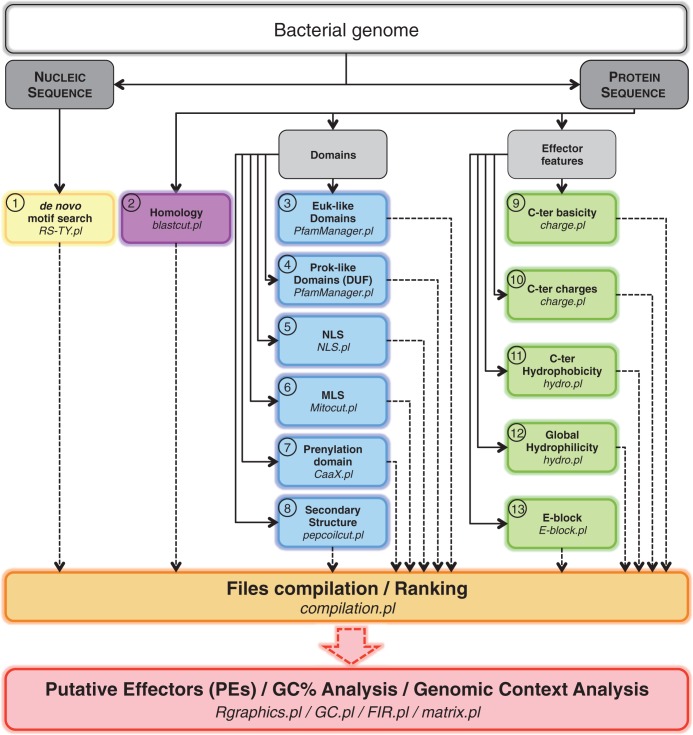
Figure 1.
Flowchart of the bioinformatics search by S4TE to identify putative effector proteins (PEs). This bioinformatics pipeline is composed of 15 steps delimited by color boxes. Steps 1–13 look for T4 effector features in a given bacterial genome. Step 14 (Files compilation/Ranking) ranks and classifies the predicted T4 effectors based on their number of features to provide the best candidates for experimental validation (Supplementary Figures S2 and S3). Step 15 analyses the genome architecture and G + C content and shows the distribution of predicted effectors. Programs used are indicated in italics. Euk-like, Eukaryotic-like; Prok-like, Prokaryotic-like; NLS, nuclear localization signal; MLS, mitochondrial localization signal; C-ter, C-terminal.