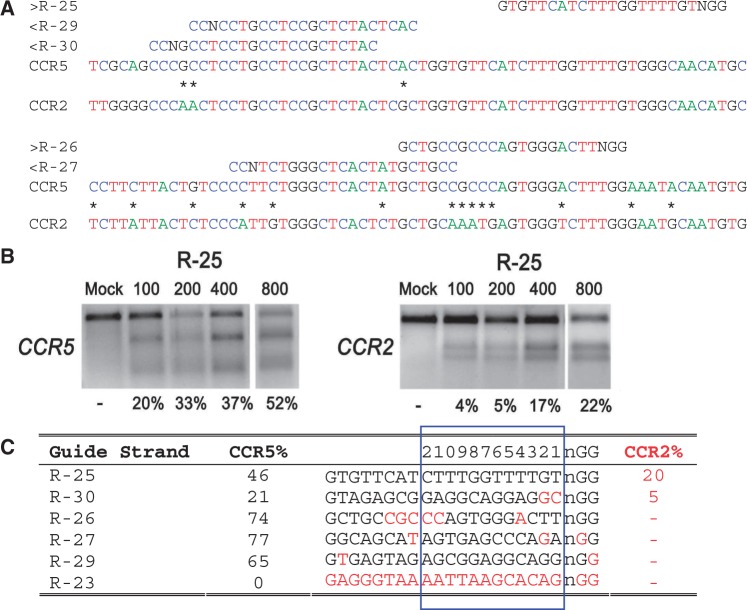
Figure 2.
On- and off-target cleavage by CRISPR/Cas9 systems targeting the CCR5 gene. (a) Guide strands are aligned to their target sites in CCR5 and corresponding region in CCR2. Forward direction guide strands (marked ‘greater than’) are shown adjacent to NGG, representing the PAM sequence. Guide strands complementary to the reverse strand (marked ‘less than’) are listed to the right of CCN. Asterisks between CCR5 and CCR2 indicate nucleotides that differentiate the two genes. For clarity, the A, C, T and G nucleotides are shown in green, blue, red and black, respectively. (b) Cells were transfected with 100, 200, 400 or 800 ng of the R-25 CRISPR plasmid. Results of the T7E1 mutation detection assay with the on- and off-target mutation rates at CCR5 and CCR2, respectively. R-23 targeting CFTR was used as a negative control. (c) Guide strands ranked in order of the off-target mutation rates at CCR2. Differences between the guide strand sequence and complementary sequence in CCR2 are in red. The 12 bases closest to the PAM are boxed in blue and numbered on top.