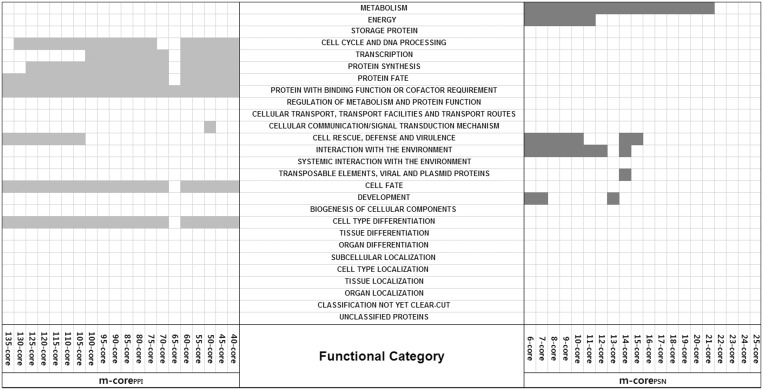
Figure 4.
The distribution of functional annotation categories (FDR-adjusted P < 0.01, see ‘Materials and Methods’ section) in each group of m-corePSN (m ≥ 6) and m-corePPI (m ≥ 40). If a functional category is significantly overrepresented in a group, then the corresponding rectangle is colored in orange for the protein interactome and in blue for the perturbation sensitivity network. The hubs of the protein interactome are enriched with ‘cell cycle and DNA processing’, ‘transcription’, ‘protein synthesis’, ‘protein fate’, ‘protein with binding function or cofactor requirement’, ‘cell fate’ and ‘cell type differentiation’, whereas those of the perturbation sensitivity network are enriched with metabolism, ‘cell rescue’, ‘defense and virulence’, ‘interaction with the environment’, ‘transposable elements’ and ‘development’. Two hubs have mutually exclusive functional annotations.