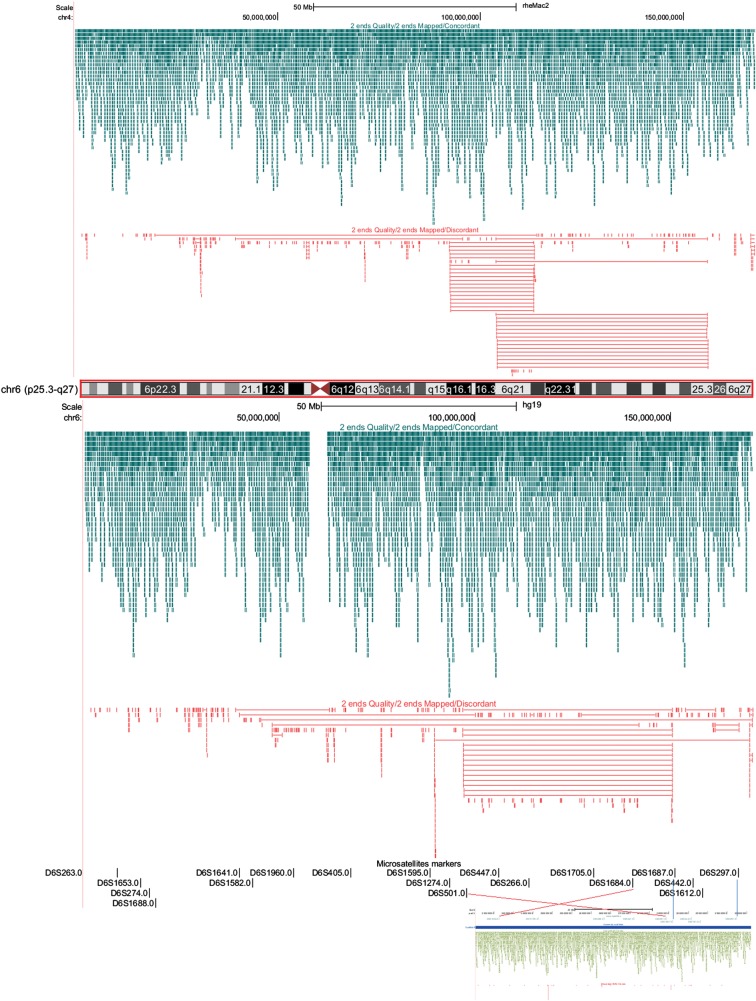
Figure 3.
Vervet genome rearrangements provide a new perspective on primate genome evolution. The panels show UCSC Genome Browser views of vervet bacterial artificial chromosome (BAC) paired ends aligned to rhesus chr6 (rheMac.2, top) human chr6 (hg19, middle), and the vervet draft assembly (bottom). Green lines delineate vervet BAC paired ends concordant for interval distance and sequence orientations. Red lines delineate vervet BAC end pairs, discordant for either interval distance, sequence orientations, or both. Clusters of concordant BACs indicate cross-species syntenic regions, whereas clusters of discordant BACs indicate rearrangements. In the example, the vervet orthologous region is organized differently from both rhesus and human. Vervet BAC end pairs aligned to a 70-Mb vervet scaffold (bottom) demonstrate that the rearrangements have been accurately reconstructed in the vervet genome assembly.