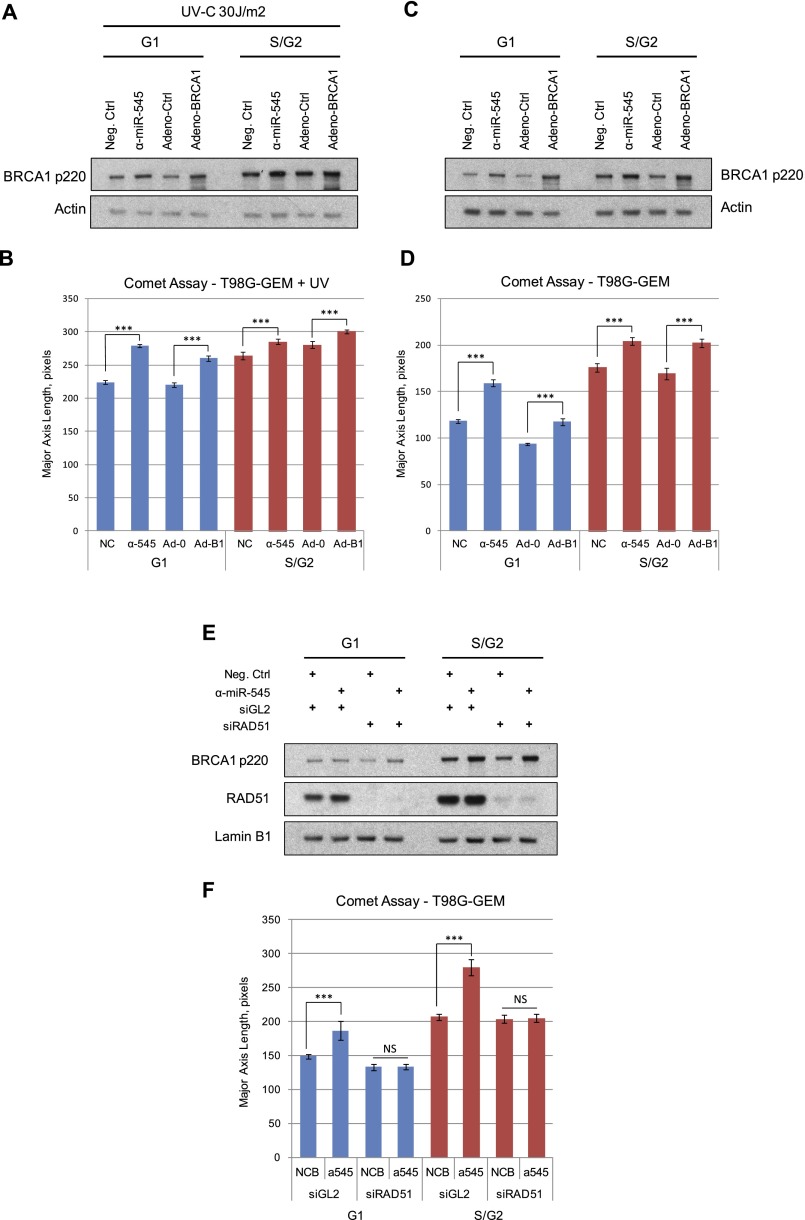
Figure 7.
Formation of DNA strand breaks in BRCA1-overexpressing G1 and S/G2 cells. Exponentially growing T98G-GEM cells were transfected with miR-545 antagomir (α-miR-545) or a negative control or infected with a recombinant BRCA1-expressing adenoviral vector (Adeno-BRCA1) or control adenoviral vector (Adeno-Ctrl). Thirty-six hours later, one set of samples was irradiated with 30 J/m2 UV-C and incubated for an additional 3 h. G1 and S/G2 subpopulations were separated by FACS from both nonirradiated and UV-irradiated samples and analyzed for the presence of DNA breaks by single-cell gel electrophoresis (comet assay). (A,C) Western blot showing the expression of BRCA1 p220 in G1 and S/G2 populations of both UV-irradiated (A) and nonirradiated (C) T98G-GEM cells. Actin served as a loading control. (B,D) Quantitation of the average tail length of comets developed in miR-545 antagomir- or negative control-treated or Adeno-BRCA1- or control adenovirus-transduced T98G-GEM G1 and S/G2 cells. (E) Western blot showing the expression of BRCA1 p220 and RAD51 in T98G-GEM cells transfected with a miR-545 antagomir or a negative control and a RAD51-specific siRNA (siRAD51) or a negative control (siGL2) as indicated, incubated for 72 h, and separated by FACS into G1 and S/G2 populations. Lamin B1 served as a loading control. (F) Quantitation of the average tail length of comets developed in the miR-545 antagomir- or negative control-treated cells transfected either with RAD51-specific siRNA or control reagent (siGL2). Comet tails were detected and analyzed by CellProfiler software (Carpenter et al. 2006). Data represent the average tail length calculated from analysis of at least 100 cells per sample in three independent experiments. Error bars indicate the SEM. Statistical significance was determined by a Student's t-test; (***) P < 0.001; (NS) nonsignificant, P > 0.05.