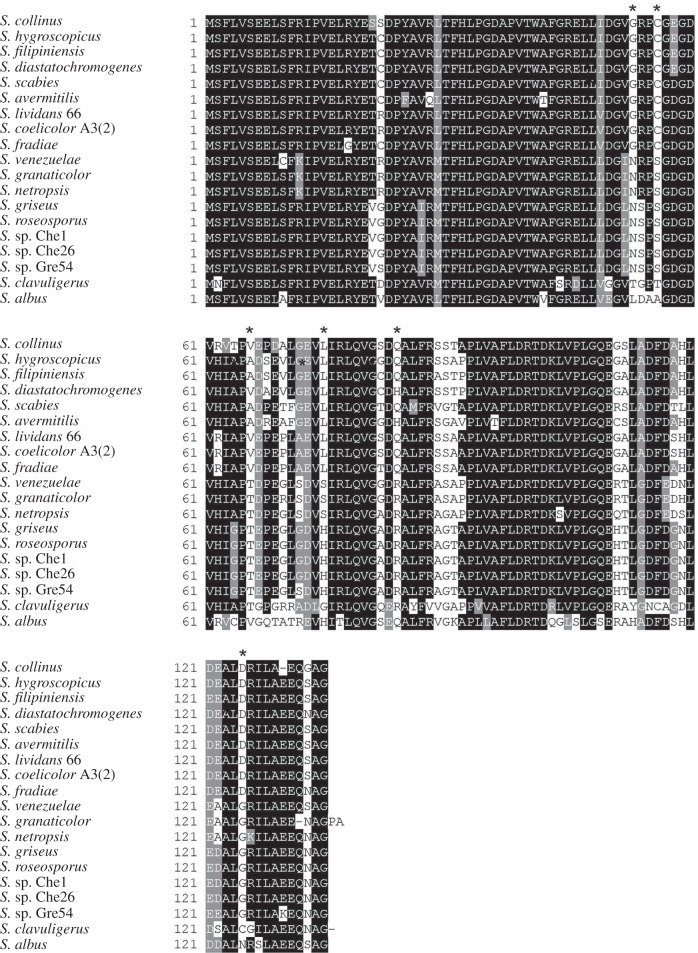
Figure 4.
Alignment of SsgA orthologues. Only those SsgA protein sequences have been used as input that are derived from species with known phenotype in submerged cultures. For shading, at least 60% of the aligned proteins should share the same or similar aa residues. Identical residues shaded black, similar residues shaded grey. Residues highlighted with an asterisk above the alignment, are conserved within—but different between—the ‘LSp’ and ‘NLSp’ branches in figure 5 and function as identifiers for the ability of a certain Streptomyces species to sporulate in submerged culture. Sequences were labelled by their strain of origin, for sequence labels see §4.2. For input sequences and their accession numbers, see the electronic supplementary material, data file S4.