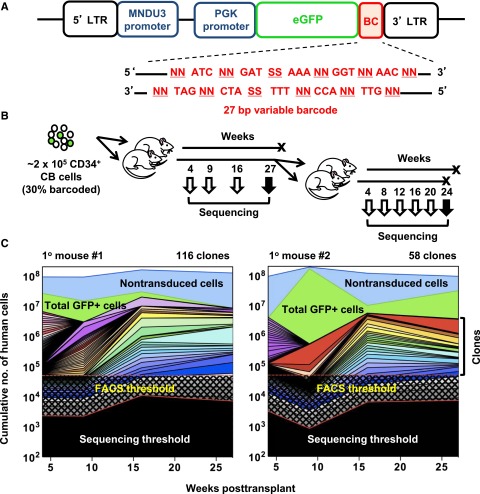
Figure 1.
Experimental design and analysis of clones detected. (A) Schematic outline of the MPG lentiviral vector that contained a 27-nucleotide non-coding DNA barcode sequence inserted downstream of the GFP reporter gene. The barcode sequence was designed with variable nucleotide doublets (NN) repeated 5 times, each separated by a constant nucleotide triplet sequence as previously described.10 (B) Schematic outline of the experimental design. CD34+ CB cells were transduced with the lentiviral barcode library for 6 hours in vitro, of which 105 cells (∼3.6 × 104 barcoded cells) were then intravenously transplanted into each of 2 sublethally irradiated NSG mice. BM aspirates were collected, total human CD45+ analyzed, and myeloid, B-, and T-cell subsets of GFP+ and GFP- cells were also separately isolated by FACS. After 27 weeks, all BM cells were harvested separately from both legs and pelvis of both primary mice. From one mouse, half the cells were sorted for clonal analysis and the other half were transplanted intravenously into 2 secondary mice. From the second primary mouse, all harvested BM cells were sorted and used for clonal analysis. (C) Analysis of clonal dynamics in the 2 transplanted primary mice. The upper blue line denotes the total number of human hematopoietic cells in the BM of each mouse (assumed to have a total cellularity of 2 × 108) and the upper green line denotes the total number of GFP+ cells in each mouse. Each color underneath denotes the total contribution of all of the lineages detected in each clone, as inferred from the BM sample analyzed. Black checked and shaded regions indicate the detection limit of the FACS (5 × 104 cells) and MPS (variable) methods, respectively.