Background: Endothelial-to-mesenchymal transition (EndMT) is involved in embryonic cardiovascular development.
Results: HDAC3 undergoes unconventional splicing during stem cell differentiation, which contributes to EndMT.
Conclusion: HDAC3 unconventional splicing may modulate endothelial cell plasticity.
Significance: Targeting HDAC3 splicing may provide new therapeutic strategies to tackle cardiovascular disease caused by endothelial plasticity.
Keywords: Cardiovascular, Endothelial Cell, Epithelial to Mesenchymal Transition, Histone Deacetylase, Transforming Growth Factor beta (TGFbeta)
Abstract
Histone deacetylase 3 (HDAC3) plays a critical role in the maintenance of endothelial integrity and other physiological processes. In this study, we demonstrated that HDAC3 undergoes unconventional splicing during stem cell differentiation. Four different splicing variants have been identified, designated as HD3α, -β, -γ, and -δ, respectively. HD3α was confirmed in stem cell differentiation by specific antibody against the sequences from intron 12. Immunofluorescence staining indicated that the HD3α isoform co-localized with CD31-positive or α-smooth muscle actin-positive cells at different developmental stages of mouse embryos. Overexpression of HD3α reprogrammed human aortic endothelial cells into mesenchymal cells featuring an endothelial-to-mesenchymal transition (EndMT) phenotype. HD3α directly interacts with HDAC3 and Akt1 and selectively activates transforming growth factor β2 (TGFβ2) secretion and cleavage. TGFβ2 functioned as an autocrine and/or paracrine EndMT factor. The HD3α-induced EndMT was both PI3K/Akt- and TGFβ2-dependent. This study provides the first evidence of the role of HDAC3 splicing in the maintenance of endothelial integrity.
Introduction
As a key cellular component of the circulatory system, endothelial cells (ECs)3 play important roles in cardiovascular homeostasis and disease development. In response to local flow patterns and physiological stimuli, ECs can exhibit a wide range of phenotypic variability throughout the cardiovascular system (1). The most remarkable feature is the plasticity of endothelial-to-mesenchymal transition (EndMT), which contributes to embryonic cardiogenesis (2–6); postnatal angiogenesis (7); and pathological processes, such as cardiac (8) and renal fibrosis (9), pulmonary hypertension (10), and tumor angiogenesis and metastasis (11, 12).
EndMT defines a specific type of cell plasticity. During EndMT, resident ECs delaminate from an organized cell layer and invade the underlying tissue. These cells lose cell-cell junctions due to decreased VE-cadherin, acquire invasive and migratory properties, and lose other EC markers, such as CD31. On the other hand, these cells gain mesenchymal markers (e.g. FSP1 (fibroblast-specific protein 1), N-cadherin, and α-smooth muscle actin (αSMA)) (5, 8, 10, 13, 14). EndMT was originally identified in cardiogenesis. A subset of ECs in the endocardium of mouse embryos at 9.5 days postcoitus (dpc) transform into mesenchymal cells that migrate, proliferate, and eventually remodel the cardiac cushions into heart valve leaflets and septa for a partitioned heart (2, 3). Recent reports revealed that EndMT accounts for up to 40% of cancer-associated fibroblasts, which is the main source of host-derived vascular endothelial cell growth factor (VEGF) (11). Several signaling pathways are reported to be involved in EndMT, such as transforming growth factor β (TGFβ) binding and Notch and Akt1 activation (5, 8, 15). A very recent study showed that EndMT is also involved in neointima formation (16). However, the mechanism of EndMT is still poorly understood.
Histone deacetylases (HDACs) modulate chromatin structure through regulating the acetylation status of histone tails, functioning as transcriptional co-repressors (17, 18). Recent studies showed that HDACs can also modulate transcription factor activity, increase gene transcription (19), and interact with cytoskeleton and signal transducers (20–22). There are 18 types of HDACs, classified into four categories. HDAC3 is a member of the Class I HDACs (17, 23). It is an indispensable gene, removal of which in the germ cell line causes embryonic lethality at an early stage (24). Our previous study indicated that HDAC3 is essential for EC differentiation and integrity maintenance (25–27). In this study, we found that HDAC3 undergoes unconventional splicing during embryonic stem (ES) cell differentiation and development. In addition, overexpression of the splicing isoform of HDAC3, HD3α, reprogrammed ECs into mesenchymal cells.
EXPERIMENTAL PROCEDURES
Materials
All cell culture media and sera were purchased from Invitrogen, and cell culture supplements and growth factors were obtained from Sigma-Aldrich. Antibodies against HD3α and -β were developed by Genescript (Piscataway, NJ) in rabbits with peptide PQGDTILTSPQNDL. The same peptide was used as a blocking peptide to test the specificity of anti-HD3α/β antibody. Antibodies against CD31 (sc-1506), N-cadherin (sc-6461), VE-cadherin (VE-Cad) (sc-9989), GAPDH (sc-25778), Akt1/2 (sc-8312), pAktS473 (sc-7985R), VEGF (sc-507), Snail2 (sc-15391), Smad3 (sc-101154), and CD9 (sc-13118) were purchased from Santa Cruz Biotechnology; antibodies against FLAG (F2426, F1804, and F7425), HA (H6908), αSMA (A5228), tubulin (T5201), HDAC3 (H3034), histone H4 (SAB4500306), pSmad2 (SAB4300251), and pSmad3 (SAB4300253) were from Sigma; and antibodies against FSP1 (ab41532), TGFβ1 (ab9758), TGFβ2 (ab10850), PI3K p85α (ab22653), and collagen IV (ab19808) were from Abcam (Cambridge, UK). Rat anti-CD31 (553369) was from BD Biosciences. All secondary antibodies were from Dakocytomation (Glostrup, Denmark). All other chemicals were purchased from Sigma.
Cell Culture
Mouse embryonic stem cells (ES-D3 cell line, CRL-1934 (ATCC, Manassas, VA)), human aortic endothelial cells (HAECs), and 293 cells were maintained as described previously (26, 28). Sca1+ cells were isolated from collagen IV-differentiated ES cells and maintained in differentiation medium (DM; α-minimum Eagle's medium supplemented with 10% FBS from Sigma, 5 × 10−5 mol/liter β-mercaptoethanol, 100 units/ml penicillin/streptomycin) for more than five passages. Because the majority of these Sca1+ cells are ECs, these are therefore designated as esECs and maintained in DM (27). For ES differentiation, the cells were seeded into a collagen IV- or I-coated flask in DM for 72 h, followed by assessments.
Plasmid Construction and Transient Transfection
HDAC3 splicing variants were amplified with a primer set from differentiated mouse ES cells and cloned into the KpnI site of pShuttle2-FLAG vector as described previously (26), verified by DNA sequencing, and designated as HD3α, -β, -γ, and -δ, respectively. For the ES or esEC differentiation assay, the cells were transfected with 2 μg/1 × 106 cells of plasmids (pShuttle-FLAG vector, pShuttle-FLAG-HDAC3, or pShuttle-FLAG-HD3α) with the nucleofection kit (Lonza, Surrey, UK) and cultured in collagen IV-coated dishes in DM in the presence of 5 ng/ml mouse VEGF (Sigma) or 20 ng/ml rat platelet-derived growth factor-BB (PDGF-BB) (Sigma) for 72 h. For co-immunoprecipitation, 293 cells were co-transfected with pShuttle-FLAG-HD3α, -β, -γ, -δ/pShuttle-HA-HDAC3 or pShuttle-FLAG-HD3α, -β, -γ, -δ/pShuttle-HA-Akt1/2 with a nucleofection kit at 2 μg/1 × 106 cells and cultured for 24 h.
Adenoviral Gene Transfer
Ad-HD3α viral vector was created from pShuttle2-FLAG-HD3α plasmid, and the resulting adenoviral particles were amplified using an adenoviral expression system (Clontech, Takara Biosciences, St Germaine-en-laye, France) and the manufacturer's protocol. For adenoviral gene transfer, HAECs were incubated with Ad-null or Ad-HD3α virus at the multiplicity of infection (MOI) indicated for 6 h. After the removal of virus solution, the cells were cultured in human endothelial serum-free medium (Invitrogen) for 24–48 h, in M199 medium supplemented with 1% FBS for 5 days, or in M199 medium supplemented with 5 ng/ml insulin for 24 h. For inhibitor or neutralization antibody assays, inhibitors (at the concentrations indicated in the figure legends) and neutralization antibody (0.2 μg/ml) were included in the whole infection and incubation process. DMSO (same volume) and IgG (0.2 μg/ml) were included as vehicle and negative control, respectively.
HD3α/β-GFP Transgenic Vector Construction
An HDAC3 DNA fragment covering exon 4 to exon 15 was amplified by PCR from genomic DNA and inserted into pLoxPneo vector. GFP coding sequences were inserted into the open reading frame of HD3α and -β within intron 12 upstream of the stop codon, whereas a LoxP-Neo-LoxP cassette was inserted into intron 12 downstream of the stop codon, creating the pLoxPneo-HD3α/β-GFPkin plasmid. ES-D3 cells were transfected with this plasmid. The positive transfection clones were selected with G418, whereas the recombinant clones were further selected with ganciclovir. The positive recombinant clones were then transfected with pCMV-Cre210 plasmid (Addgene, Cambridge, MA) to remove the loxP-Neo-LoxP cassette. The positive HD3α/β-GFP stable cell clones were verified by PCR with primer sets flanking the GFP insertion and LoxP site, respectively. For GFP observation, HD3α/β-GFP-transfected ES cells were cultured in collagen I-coated dishes in DM for 3 days and observed under inverted phase and fluorescence microscopes. Images were taken and processed by Photoshop software.
Indirect Immunofluorescence Staining
Cryosections (5 μm) from mouse embryos at 10.5- and 12.5-dpc stages or chamber slides of cell culture were fixed with methanol at room temperature for 15 min, permeabilized with 0.1% Triton X-100/PBS for 15 min, blocked with normal donkey serum (1:20 dilution in PBS) for 1 h, and incubated with primary antibodies (diluted in blocking serum) and Alexa Fluor®546/488-conjugated secondary antibodies from donkeys (Dako) at 37 °C for 1 h and 30 min, respectively. Nuclei were counterstained with DAPI. To assess the specificity of HD3α antibody, 10 μg/ml HD3α antibody was incubated with a 10 μg/ml concentration of the HD3α peptide (used to raise the antibody) at 37 °C for 1 h prior to primary antibody incubation. Images were obtained with a Leica SP5 confocal microscope and LASAF software (Leica) at 37 °C and processed by Adobe Photoshop software.
Mouse Embryonic and Adult Organ Tissues
Pregnant C57BL/6J female mice were sacrificed, and the whole embryos at 8.5, 10.5, 13.5, 15.5, 17.5, and 19.5 dpc and in new born mice at day 1 were harvested for RNA extraction. Meanwhile, different organ tissues were harvested from the female mice for RNA extraction. All animal experiments in this study were performed according to protocols approved by the Institutional Committee for the Use and Care of Laboratory Animals.
Quantitative RT-PCR Assay
Cellular total RNA was isolated using the RNeasy kit (Qiagen, Manchester, UK) based on the manufacturer's protocol. One μg of RNA was reverse transcribed into cDNA with the ImproRT system (Promega, Southampton, UK) using random primer, and 10 ng of cDNA (relative to the RNA amount) was used in quantitative PCR with the SYBR Green kit (Invitrogen) to analyze the expression of mouse HDAC3 (5′-tatggctgagacaccagagtg-3′ and 5′-atctggtccagatactgggtgag-3′), VE-Cad (5′-atctgtgccagagatgtcagc-3′ and 5′-gaatgtgtactgctggtagac-3′), and SM22 (5′-catgagccgagaagtgcactc-3′ and 5′-ctaagcaggatgctgcagctc-3′) and human TGFβ1 (5′-atcctgcatctggtcacggtc-3′ and 5′-cttggcgtagtactcttcgtc-3′), TGFβ2 (5′-aagactatcgacatggagctg-3′ and 5′-gtaccgcttctcggagctctg-3′), TGFβ3 (5′-gcacaacgaactggctgtctg-3′ and 5′-aacagccactcacgcacagtg-3′), αSMA (5′-agccaagcactgtcaggaat-3′ and 5′-caccatcaccccctgatgtc-3′), and HD3α (5′-tatggctgagacaccagagtg-3′ and 5′-ttgactcatagcctagtcctg-3′). β-actin (5′-cacaactgggacgacatggag-3′ and 5′-ttcatgaggtagtcagtctgg-3′) was included as an internal control.
Immunoprecipitation and Immunoblotting
Cells were lysed by incubation with IP-A buffer (0.02 mol/liter Tris-HCl, pH 7.5, 0.12 mol/liter NaCl, 1 × 10−3 mol/liter EDTA, 1% Triton X-100 plus protease inhibitors (Roche Applied Science)) on ice for 45 min, followed by protein concentration assessment with Bradford reagent (Bio-Rad). One mg of cell lysate was mixed with 2 volumes of IP-B buffer (IP-A without Triton X-100), precleared with 2 μg of normal IgG and 10 μl of Easyview Protein G-agarose beads (Sigma), and then incubated with 2 μg of anti-HA or anti-FLAG antibody and 10 μl Easyview Protein G-agarose beads. The immunoprecipitates were separated by SDS- PAGE and detected by Western blot analysis. Fifty μg of cell lysate was included as an input control. Immunoblotting was performed as a standard procedure described elsewhere.
Cellular Fractionation
HAECs were collected by scraping in a 400 μl/75-ml flask of hypotension buffer (0.01 mol/liter Tris-Cl, pH 7.5, 0.01 mol/liter KCl plus protease inhibitors) and incubated on ice with vortexing every 5 min for 15 min. Twenty-five μl of 10% Nonidet P-40 was added and vortexed at 200 rpm for 10 s. Nuclei were spun down at 16,100 × g at 4 °C for 10 s. The supernatant was recovered as a cytosol fraction. The nuclei were washed once with PBS, resuspended in 70 μl of hypotension buffer containing 0.625% Nonidet P-40, and sonicated for 6 s. Nuclear extract was recovered from the supernatant by spinning at 16,100 × g at 4 °C for 5 min. Protein concentration was assessed with Bio-Rad reagent. Twenty-five μg of proteins was applied to Western blot analysis.
Analysis of Secreted Proteins
HAECs were infected with Ad-null or Ad-HD3α at an MOI of 10 in the absence or presence of inhibitors for 6 h in complete growth medium, followed by washing with serum-free M199 medium three times and incubation with M199 medium supplemented with 5 ng/ml insulin for 24 h. Conditioned medium was collected, and the cell debris was removed by centrifugation at 4000 rpm at 4 °C for 5 min. The supernatant was recovered and subjected to exosome isolation and whole medium concentration, respectively. For isolation of exosomes, 1 ml of the supernatant was mixed with 500 μl of total exosome isolation reagents (Invitrogen) and incubated on a rotator at 4 °C for 24 h, followed by spinning at 16,100 × g at 4 °C for 1 h. The pellet was resuspended in 25 μl of 1× SDS loading buffer (0.02 mol/liter Tris-HCl, pH 8.9, 2% SDS, 10% glycerol, 0.5% 2-mercaptoethanol, 0.025% bromphenol blue). For total medium concentration, 500 μl of the supernatant was applied to an Amicon Ultra-0.5 Ultracel-3 membrane unit (Millipore, Watford, UK) and centrifuged at 16,100 × g at 4 °C for 15 min. This procedure was repeated until 5 ml of the supernatant was concentrated to 100 μl, which was mixed with 25 μl of 5× SDS loading buffer. Twenty-five μl of exosomes or concentration sample was applied to SDS-PAGE and Western blot analysis.
siRNA Transfection Assay
HAECs were seeded in gelatin-coated 75-ml flasks at 50% confluence 24 h before transfection and washed three times with PBS and incubated with both serum- and antibiotic-free M199 medium 1 h prior to transfection. Upon transfection, 60 μl of 1 × 10−5 mol/liter PI3K p85 siRNA (sc-39126, Santa Cruz Biotechnology, Inc.), Akt1 siRNA (sc-29196), or control siRNA (sc-37007) was diluted into 800 μl of M199 and incubated with prediluted Lipofectamine RNAiMAX (50 μl in 800 μl of M199 for 5 min; Invitrogen) at room temperature for 15 min. The mixture was added to the pretreated HAECs (6 ml/75-ml flask of M199 for 1 h) and incubated for 5 h, after which the medium was refreshed with complete growth medium. Forty-eight h post-transfection, the cells were infected with Ad-null or Ad-HD3α and cultured for 24 h, followed by incubation in serum-free M199 containing 5 ng/ml insulin for 24 h. Cell lysates and culture medium were collected for Western blot analysis of αSMA expression and TGFβ2 secretion, respectively.
Statistical Analysis
Data expressed as the mean ± S.E. were analyzed with a two-tailed Student's t test for two groups or pair-wise comparisons or analysis of variance for more than one comparison. A value of p < 0.05 was considered to be significant.
RESULTS
HDAC3 Undergoes Unconventional Splicing during Stem Cell Differentiation
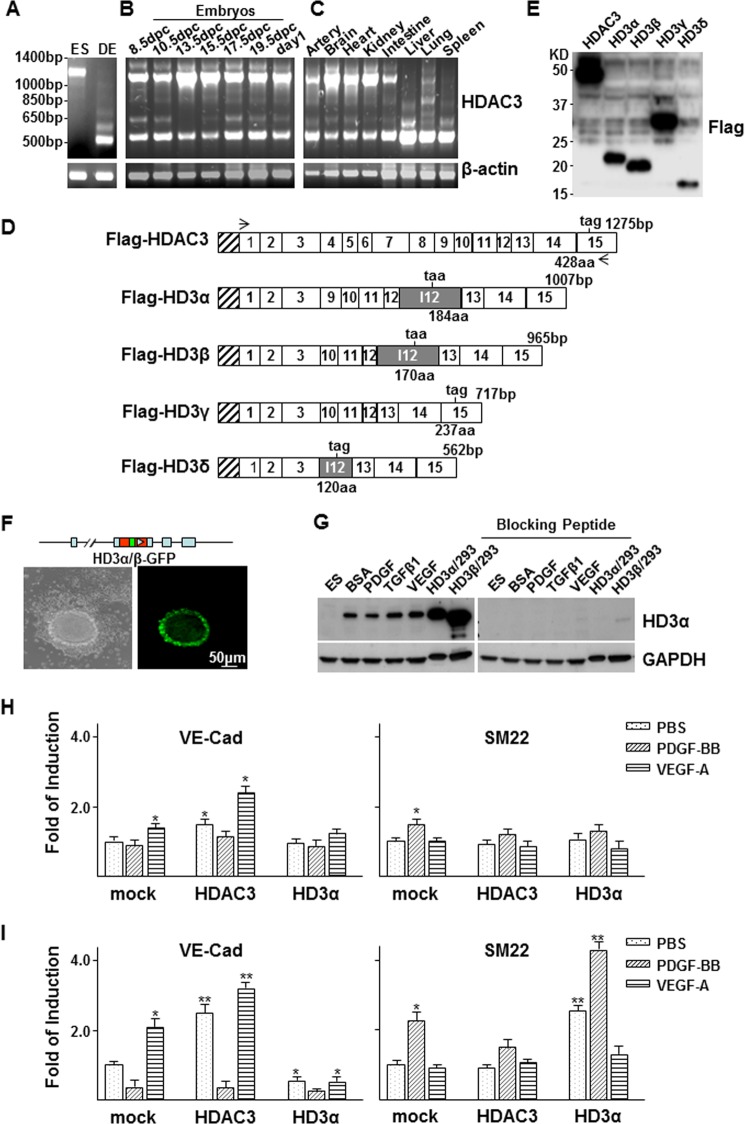
Our previous study demonstrated that HDAC3 was essential for EC differentiation (26, 27). When cloning mouse HDAC3 cDNA sequence using a primer set 5′-atgaccggtaccgtggcgtatttctacgac-3′ and 5′-cacagcaagcttgctgctctaaatctccac-3′, we discovered a few smaller electrophoretic bands in differentiated embryonic stem (DES) cell samples (Fig. 1A). Similar bands were also found in samples from mouse embryos (Fig. 1B) and adult tissues (Fig. 1C). To verify the nature of these bands, all of the bands were isolated and sequenced (GenBankTM accession numbers JN651901–JN651904). Interestingly, the bands appeared to be isoforms spliced in an unconventional way. Four different isoforms have been identified and designated as HD3α, -β, -γ, and -δ (Fig. 1D). In HD3α, -β, and -δ, intron 12 was kept as an additional exon, in which part of the intron sequence was incorporated into the open reading frame (ORF) and ended within the intron (GenBankTM accession numbers JN651901, JN651902, and JN651904). HD3α and HD3β have the same ORF, and the only difference is that HD3β loses sequence from exons 9 and 10. The ORF of HD3δ downstream exon 3 is totally different from that of HD3α and HD3β. Although all three isoforms contain sequences from intron 12, the C terminus of HD3δ is different from that of HD3α and HD3β. In addition, recombination appears to be present between exons 3 and 9 for HD3α, between exons 3 and 10 for HD3β and HD3γ, and between exon 3 and intron 12 for HD3δ. All four isoforms share the same far N-terminal domain as full-length HDAC3 and possess a specific C-terminal domain derived from intron 12, except for HD3γ. FLAG tag expression of the isoforms in 293 cells revealed the expected protein bands (Fig. 1E). To verify that these isoforms were not artifacts derived from RT-PCR, an HDAC3 gene construct was created (pLoxPneo-HD3α/β-GFPkin), in which a GFP coding sequence was inserted into intron 12 within the ORF of HD3α/β upstream of the stop codon. The construct was transfected into mouse ES cells and selected sequentially with Geneticin G-418 and ganciclovir. The HD3α/β-GFP knock-in ES cell line was eventually established via removal of the LoxP-Neo-LoxP cassette using pCMV-Cre transfection. Normally, intron 12 is removed through conventional splicing, so GFP is absent. Although HD3α/β-GFP knock-in ES cells did not yield GFP-positive cells in ES culture medium, when these cells were cultured in differentiation medium for 3 days, a portion of the cell population became GFP-positive cells (Fig. 1F), suggesting that the intron 12 was kept as an additional exon, and the intron sequence was incorporated into the ORF. These results suggest that HDAC3 naturally undergoes unconventional splicing during stem cell differentiation, as far as the HD3α/β isoforms are concerned. Because the ORF is different, HD3δ cannot give rise to GFP. To distinguish HD3α/β isoforms, we raised antibody in rabbit against peptide PQGDTILTSPQNDL from intron 12 (GenBankTM accession numbers JN651901 and JN651902) and performed Western blot on differentiated ES cell samples. A band corresponding to HD3α was detected in DES cells but not in undifferentiated ES cells (Fig. 1G, left), which disappeared when the antibody was preincubated with the blocking peptide (Fig. 1G, right), suggesting that the antibody is specific. The intensity of this HD3α band was slightly changed in PDGF-, TGFβ1-, or VEGF-treated DES cells. No band corresponding to HD3β was detected (Fig. 1G, left). Because HD3α is the main isoform, the following study will only focus on HD3α.
FIGURE 1.
HDAC3 undergoes unconventional splicing. A–C, multiple bands of HDAC3 were detected in DES cells (A), mouse embryos (B), and adult tissues (C). Undifferentiated ES cells (ES) were included as control cells, and β-actin was included as a loading control. D, schematic illustration of the sequence structure of cloned HDAC3 and splicing variants. Open boxes, exons with numbers inside; shadowed box, intron 12. The length of each isoform is indicated at the DNA base (bp) and amino acid (aa) levels. The taa or tag is the stop codon. The stretched box indicates the FLAG tag. Arrows, primer positions. E, Western blot shows the protein bands for HDAC3 and the splicing isoforms in transfected 293 cells. F and G, HDAC3 splicing occurred naturally during stem cell differentiation as revealed by GFP observation (F) and Western blot (G). F, HD3α/β-GFP knock-in (top schematic illustration) ES cells were cultured in a collagen I-coated flask in differentiation medium for 3 days, followed by imaging at inverted phase (left) and fluorescence (right) microscopes. G, ES cells were cultured in a collagen I-coated flask in differentiation medium for 2 days, followed by treatment with 10 ng/ml rat PDGF-BB, 5 ng/ml TGFβ1, or 5 ng/ml VEGF. Western blot analysis was performed with anti-HD3α antibody pretreated with (right) or without (left) blocking peptide. Cell lysates from 293 cells transfected with pShuttle2-FLAG-HD3α (HD3α/293) and pShuttle2-FLAG-HD3β (HD3β/293) were included as positive control. H and I, quantitative RT-PCR analysis of the effect of overexpression of HD3α on VE-Cad and SM22 expression in ES (E) and ES-derived EC (F) differentiation. *, p < 0.05; **, p < 0.01. Data presented are representative of or the average of three independent experiments.
Because HDAC3 is essential for EC differentiation (26), we tested whether the splicing event will affect such process by introducing HDAC3 and HD3α into DES cells and esECs (27) via plasmid transfection, followed by quantitative RT-PCR analysis of VE-Cad and SM22 gene expression. As expected, overexpression of HDAC3 increased VE-Cad expression in both ES cells (Fig. 1H, left) and esECs (Fig. 1I, left), with VEGF having a synergistic effect. HD3α had no effect on VE-Cad or SM22 gene expression in ES cells (Fig. 1H). In esECs, however, HD3α decreased VE-Cad (Fig. 1I, left) whereas it increased SM22 (Fig. 1I, right) expression, especially under PDGF treatment. These results suggest that HD3α may promote ECs trans-differentiation into mesenchymal cells.
HDAC3 Splicing May Play a Role in Embryonic Development
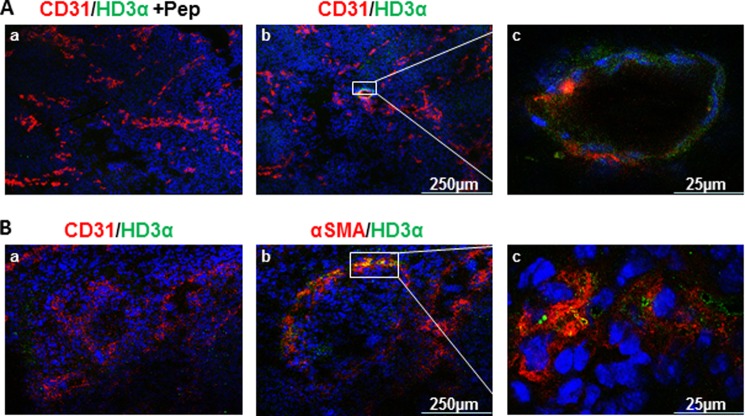
As described in the legend to Fig. 1B, spliced HDAC3 bands could be detected in mouse embryos by routine RT-PCR; it is worth investigating whether such a splicing event was involved in mouse embryonic development. Mouse embryos were harvested at 10.5 and 12.5 dpc, respectively. Cryosections were made to include the two developmental stages of the vasculogenesis, which starts from endothelial tube formation (Fig. 2A) and ends with smooth muscle cell localization (Fig. 2B). Immunofluorescence staining with anti-HD3α revealed that HD3α emerged in the CD31-positive endothelial cells during the 10.5-dpc stage when vasculogenesis begins (Fig. 2A, b and c). At 12.5 dpc, there is no colocalization of CD31 and HD3a (Fig. 2B, a). However, HD3α was found to co-localize with αSMA-positive smooth muscle cells (Fig. 2B, b and c). Preincubation with the blocking peptide abolished HD3α staining (Fig. 2A, a), confirming the specificity of anti-HD3α antibody. These results indicate that HD3α is involved in the embryonic vasculogenesis and may be a key regulator in the determination of vascular progenitor cells.
FIGURE 2.
HDAC3 splicing may contribute to mouse embryonic development. A, HD3α co-localized with CD31 at the 10.5-dpc stage. Triple staining for DAPI (blue), CD31 (red), and HD3α (green) was performed on mouse 10.5-dpc sections in the presence (a) or absence of HD3α blocking peptide. c, higher magnification shows the co-localization of HD3α and CD31. B, HD3α co-localized with αSMA (b) but not with CD31 (a) at the 12.5-dpc stage. Triple staining for DAPI (blue), CD31 (red; a)/αSMA (red; b), and HD3α (green) was performed on mouse 12.5-dpc sections. c, higher magnification shows the co-localization of HD3α and αSMA. Data shown are representative of at least three independent experiments.
Overexpression of HD3α Induces EndMT via PI3K-Akt and TGFβ Signal Pathways
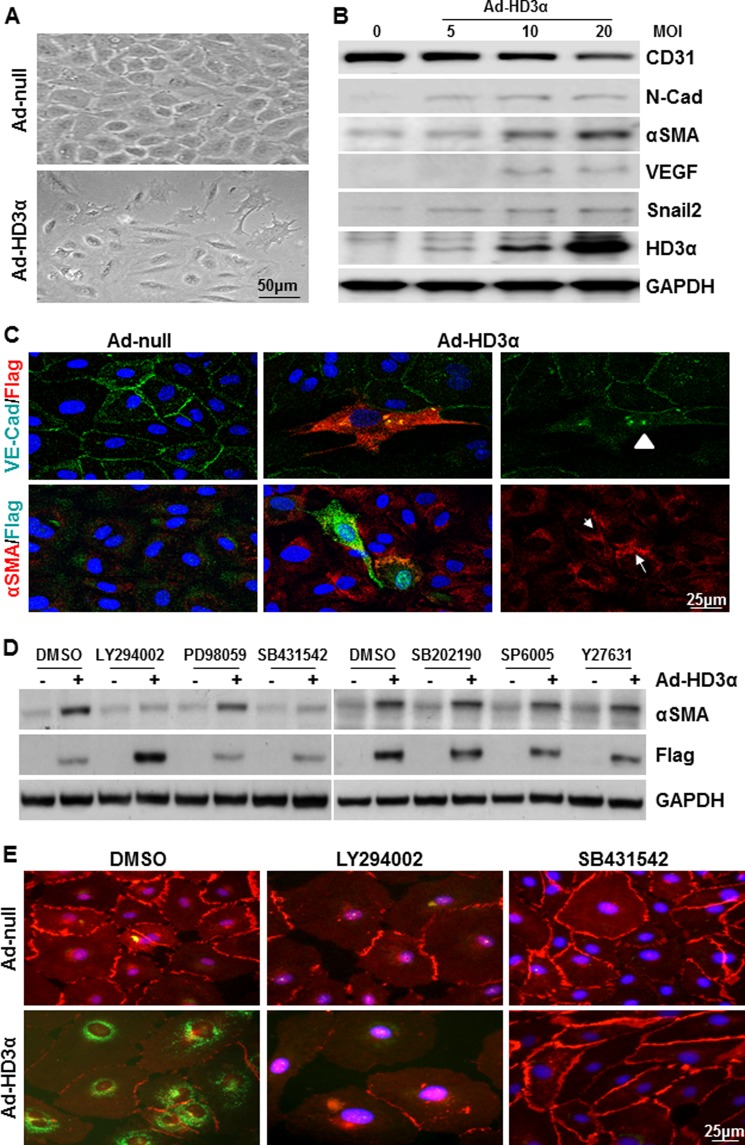
To confirm whether HD3α is involved in EndMT, the effect of overexpression of HD3α in mature endothelial cell was investigated. HAECs were infected with Ad-HD3α virus followed by incubation with serum-free culture medium or M199 medium supplemented with 1% FBS. 24 h postinfection, the cell growth rate decreased, and some cells underwent morphology change (Fig. 3A). Western blot analysis revealed that overexpression of HD3α decreased the EC marker CD31, whereas it increased mesenchymal markers, such as N-cadherin, αSMA, VEGF, and EndMT transcription factor Snail2, in a dose-dependent manner (Fig. 3B). Immunofluorescence staining confirmed the EndMT phenotype as revealed by the decrease or redistribution of VE-Cad and the occurrence of αSMA in the Ad-HD3α-infected and adjacent cells (Fig. 3C). These results suggest that overexpression of HD3α can induce EndMT.
FIGURE 3.
Overexpression of HD3α induces EndMT via PI3K/Akt and TGFβ signal pathways. A, overexpression of HD3α induced cell morphology change in HAECs. Images were taken at day 5 postinfection. B, overexpression of HD3α induced the EndMT phenotype in a dose-dependent manner as revealed by the decrease of CD31 and the increase of mesenchymal markers. C, immunofluorescence staining revealed that overexpression of HD3α induced the redistribution of VE-Cad (green) in HD3α-positive cells (FLAG; red, arrowhead) and the expression of αSMA (red) in HD3α-positive cells (FLAG; green, arrow) and adjacent cells. D, PI3K-Akt and TGFβ signal pathways were essential for HD3α-induced αSMA expression in HAECs. Inhibitors LY294002 (5 × 10−6 mol/liter), PD98059 (1 × 10−5 mol/liter), SB431542 (1 × 10−5 mol/liter), SB202190 (1 × 10−5 mol/liter), SP6005 (1 × 10−5 mol/liter), and Y27632 (1 × 10−5 mol/liter) were included in the virus infection (10 MOI, 6 h) and incubation in serum-free medium (24 h). Ad-null was included as virus control, and DMSO was included as vehicle control. FLAG antibody was used to detect exogenous HD3α. E, both LY294002 and SB431542 abolished Ad-HD3α-induced EndMT. 5 × 10−6 mol/liter LY294002 or 1 × 10−5 mol/liter SB431542 was included in the virus infection (10 MOI for 6 h) and further incubation in M199 medium supplemented with 1% FBS. Immunofluorescence staining was performed to detect VE-Cad (red) and αSMA (green) at day 5 postinfection. DMSO was included as a vehicle control. Data presented are representative of three independent experiments.
Multiple signal pathways have been reported to be involved in the EndMT process, such as TGFβ, PI3K/Akt, MAPK, p38, JNK, RhoA, etc. (8, 15, 29–33). To identify which signal pathway is involved in HD3α-induced EndMT, specific inhibitors LY294002 (PI3K/Akt), PD98059 (MAPK), SB202190 (p38), SB431542 (TGFβ), SP60005 (JNK), and Y27631 (RhoA) were tested. As shown in Fig. 3D, both LY294002 and SB431542 abolished Ad-HD3α-induced αSMA expression in HAECs, whereas the other inhibitors did not. The inhibitory effect of LY294002 and SB431542 was further confirmed by immunofluorescence staining with anti-VE-Cad (red) and anti-αSMA (green) antibodies (Fig. 3E). These experiments suggest that both PI3K/Akt and TGFβ signal pathways are required by HD3α in driving EndMT.
HD3α Induces TGFβ2 Activation
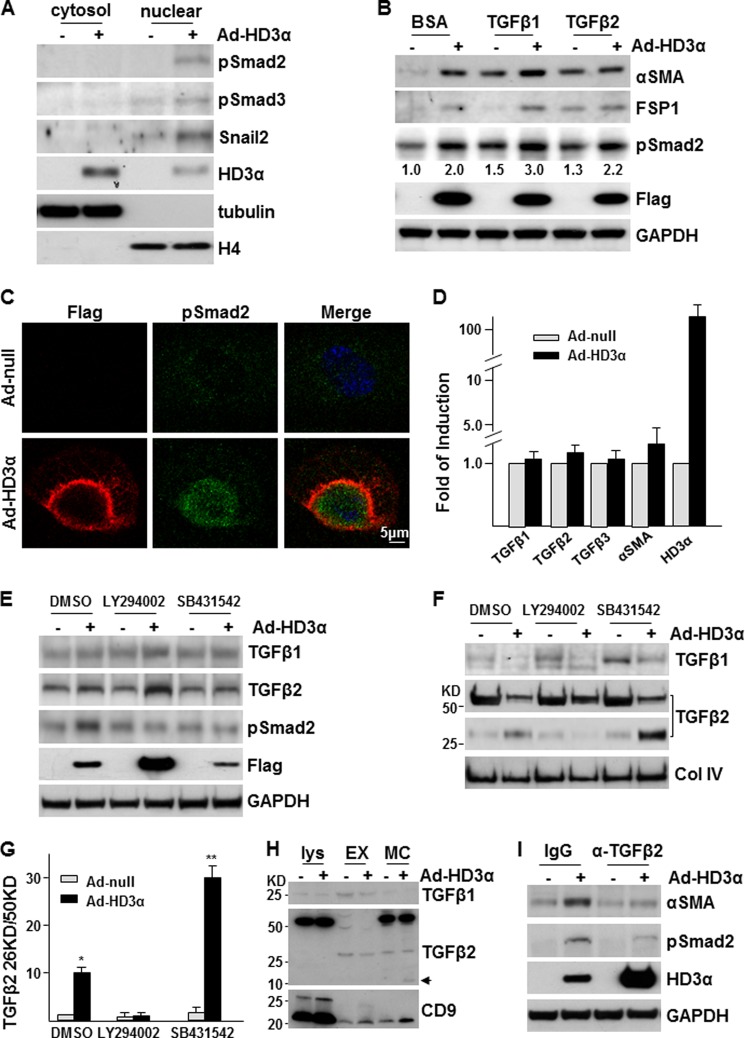
The TGFβ signaling pathway plays a critical role in EndMT (34). The above study has demonstrated that this pathway is also required by HD3α-induced EndMT. To investigate how HD3α drives EndMT through TGFβ signal pathway, the activation of this pathway was first assessed in Ad-HD3α-infected cells. As shown in Fig. 4A, overexpression of HD3α induced Smad2/3 phosphorylation and nuclear translocation and increased Snail2 expression and nuclear translocation. The majority of HD3α itself is located in the cytosol, with only a small portion located in the nuclear fraction. Further experiments indicated that exogenous TGFβ1 and -2 could induce EndMT, but only TGFβ1 had an additive effect on HD3α-induced EndMT (Fig. 4B). In the presence of TGFβ1, overexpression of HD3α induced a huge accumulation of phospho-Smad2 in the nuclei, whereas the majority of HD3α is located on the nuclear envelope (Fig. 4C). These experiments suggest that HD3α drives EndMT through activating Smad2/3 phosphorylation and nuclear translocation.
FIGURE 4.
Overexpression of HD3α induces EndMT through TGFβ2 secretion and activation. A, overexpression of HD3α activated the TGFβ signaling pathway. Tubulin and histone H4 were used to indicate cytosol and nuclear extract fractions, respectively. B, TGFβ1 enhanced HD3α-induced EndMT. Ad-null- or Ad-HD3α-infected HAECs were treated with 5 ng/ml TGFβ1 or TGFβ2 in serum-free medium for 24 h, followed by Western blot analysis. Numbers are relative amounts of phospho-Smad2 averaged from three independent experiments. C, overexpression of HD3α enhanced TGFβ1-induced Smad2 phosphorylation (green) and nuclear translocation. FLAG antibody was used to identify exogenous HD3α (red). D, overexpression of HD3α did not affect TGFβs mRNA levels. E–G, LY294002 abolished Ad-HD3α-induced TGFβ2 activation and Smad2 phosphorylation. 5 × 10−6 mol/liter LY294002 or 1 × 10−5 mol/liter SB431542 was included in the virus infection (10 MOI for 6 h) and incubation process (M199 supplemented with 5 ng/ml insulin for 24 h), followed by Western blot analysis (without 2-mercaptoethanol) of cell lysates (E) and conditioned media (F and G). DMSO was included as a vehicle control. *, p < 0.05; **, p < 0.01. H, TGFβ2 did not exist in exosomes. Ad-null or Ad-HD3α-infected HAECs were incubated with serum-free medium containing 5 ng/ml insulin for 24 h, followed by Western blot analysis (with 2-mercaptoethanol) of cell lysate (Lys; 25 μg of protein), exosomes (EM; from 1 ml of medium/lane), and whole medium concentrates (MC; from 1 ml/lane). CD9 was used to indicate exosomes. Arrow, the 13 kDa active band. I, TGFβ2 neutralization antibody attenuated HD3α-induced Smad2 phosphorylation and αSMA expression. For all Western blot analyses, FLAG antibody was used to detect exogenous HD3α, whereas GAPDH was included as a loading control. Data are representative of or the average of three independent experiments.
In the TGFβ signaling axis, TGFβs bind to TGFβ receptor-II, which recruits and phosphorylates TGFβ receptor-I (ALK5). The latter activates Smad2/3 phosphorylation and nuclear translocation (35). As described above (Fig. 3, D and E), ALK5 is essential for HD3α-induced EndMT. Furthermore, adjacent cells next to Ad-HD3α-infected cells also express αSMA (Fig. 3C). It is reasonable to assume that HD3α induces the secretion and/or activation of TGFβs, which in turn activate ALK5 in an autocrine and paracrine manner. To test this, we first detected TGFβ1, -2, and -3 mRNA levels by quantitative RT-PCR. Overexpression of HD3α did not seem to affect these gene expressions at mRNA levels (Fig. 4D). Interestingly, there was also only a slight increase at the αSMA mRNA level (Fig. 4D). Increase of αSMA protein may be mediated by translational regulation. Western blot analysis showed that overexpression of HD3α had no significant effect on TGFβ1 and TGFβ2 protein levels in cell lysate as well, although Smad2 phosphorylation was significantly increased (Fig. 4E). However, the analysis of the secreted proteins in conditioned medium indicated that overexpression of HD3α significantly increased TGFβ2 activation via cleavage, as revealed by the occurrence of the small electrophoretic band (Fig. 4, F and G). PI3K-Akt inhibitor, LY294002, increased TGFβ1 and TGFβ2 protein levels in Ad-HD3α-infected cells but abolished Ad-HD3α-induced Smad2 phosphorylation (Fig. 4E) and attenuated TGFβ2 activation (Fig. 4, F and G). As expected, ALK5 inhibitor SB431542 abolished HD3α-induced Smad2 phosphorylation (Fig. 4E), and the active TGFβ2 was accumulated in Ad-HD3α-infected cell medium (Fig. 4, F and G). TGFβ1 is reported to be secreted by exosomes (36). To test whether the secretion of TGFβ2 is also mediated by exosomes, TGFβ2 was detected in cell lysate, exosomes, and whole medium concentrates by Western blot. Exosomes were confirmed by the presence of CD9 protein (37). As expected, a higher level of TGFβ1 was detected in exosomes as compared with the medium (Fig. 4H). However, the majority of the secreted TGFβ2, either the precursor (50 kDa band) or active band (13 kDa; arrow in Fig. 4H) locates in the whole medium concentrates but not in exosomes, suggesting that TGFβ2 is secreted by mechanisms other than exosomes. Further experiments revealed that the presence of TGFβ2 neutralization antibody attenuated HD3α-induced Smad2 phosphorylation and αSMA expression (Fig. 4I). These results suggest that HD3α induces TGFβ2 protein secretion and activation in a PI3K-Akt pathway-dependent manner and that the active TGFβ2 in turn functions as an autocrine and/or paracrine factor to activate downstream signal pathways, leading to EndMT.
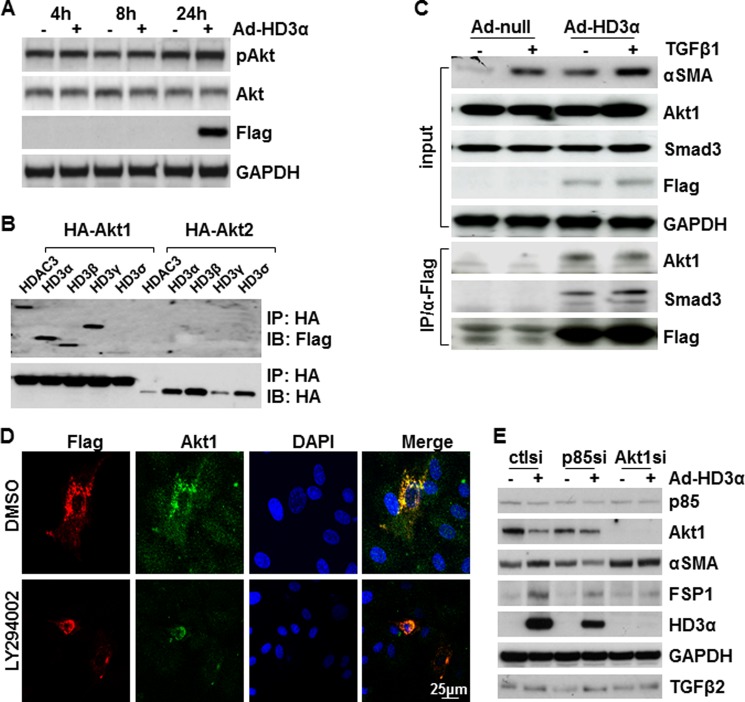
HD3α Physically Interacts with Akt1
PI3K-Akt pathway plays a central role in EMT (38). As described above, our study has demonstrated that an active PI3K-Akt pathway is essential to HD3α-induced TGFβ2 activation and EndMT. The question is how PI3K-Akt cross-talks with HD3α. To test this, Akt phosphorylation was assessed by Western blot following Ad-HD3α infection. As shown in Fig. 5A, overexpression of HD3α did not cause a significant up-regulation of Akt phosphorylation, suggesting that HD3α itself does not activate the PI3K-Akt pathway. Our previous study has shown that HDAC3 could associate with Akt1 (25). We tested whether HD3α could also physically interact with Akt by co-transfecting FLAG-tagged HDAC3 and isoform plasmids together with HA-tagged Akt1 or Akt2 into 293 cells, followed by co-immunoprecipitation assays. HDAC3 and all four isoforms associated with Akt1 but not with Akt2 (Fig. 5B). Further experiments revealed that HD3α associated with endogenous Akt1 in HAECs and that this association was not affected by TGFβ1 (Fig. 5C). Immunofluorescence staining confirmed the association of Akt1 and HD3α in HAECs, which did not seem to be affected by the presence of LY294002 (Fig. 5D). Because LY294002 is also reported to inhibit other signal pathways like Adamts1 (39), the direct involvement of the PI3K/Akt pathway in HD3α-driven EndMT was confirmed by siRNA-mediated knockdown assays. As shown in Fig. 5E, knockdown of either PI3K p85α or Akt1 ablated Ad-HD3α-induced αSMA expression. However, HD3α-induced TGFβ2 secretion and cleavage seemed not to be affected. The deficiency of p85 and especially Akt1 significantly reduced the HD3α protein level. These results suggest that PI3K and Akt1 may not be directly involved in TGFβ2 secretion and cleavage but are essential for HD3α-induced αSMA expression.
FIGURE 5.
HD3α physically interacts with Akt1. A, HD3α did not activate Akt phosphorylation. FLAG antibody was used to detect exogenous HD3α, and GAPDH was included as a loading control. B, Western blot detected the association of HDAC3 and its isoforms with Akt1. 293 cells were transfected with FLAG-HDAC3 and FLAG-HD3α,β,γ,δ together with HA-Akt1 or HA-Akt2, followed by immunoprecipitation (IP) with anti-HA antibody and Western blot (IB) with anti-FLAG and HA antibodies. C, TGFβ1 enhanced HD3α association with Smad3 but did not affect HD3α association with Akt1. D, LY294002 did not affect the association of HD3α with Akt1. HAECs were infected with Ad-HD3α at an MOI of 10 in the presence of DMSO or 5 × 10−6 mol/liter LY294002 for 6 h and then cultured in serum-free medium in the presence of DMSO or LY294002 for 24 h, followed by immunofluorescence staining with anti-Akt1 (green) and FLAG (red) antibodies. E, PI3K and Akt1 were essential for HD3α-induced αSMA expression. HAECs were transfected with p85 siRNA or Akt1 siRNA for 48 h, followed by Ad-HD3α infection for 48 h. Prior to harvesting cell lysate and culture medium, the cells were treated with serum-free M199 for 24 h. Data presented are representative of three independent experiments.
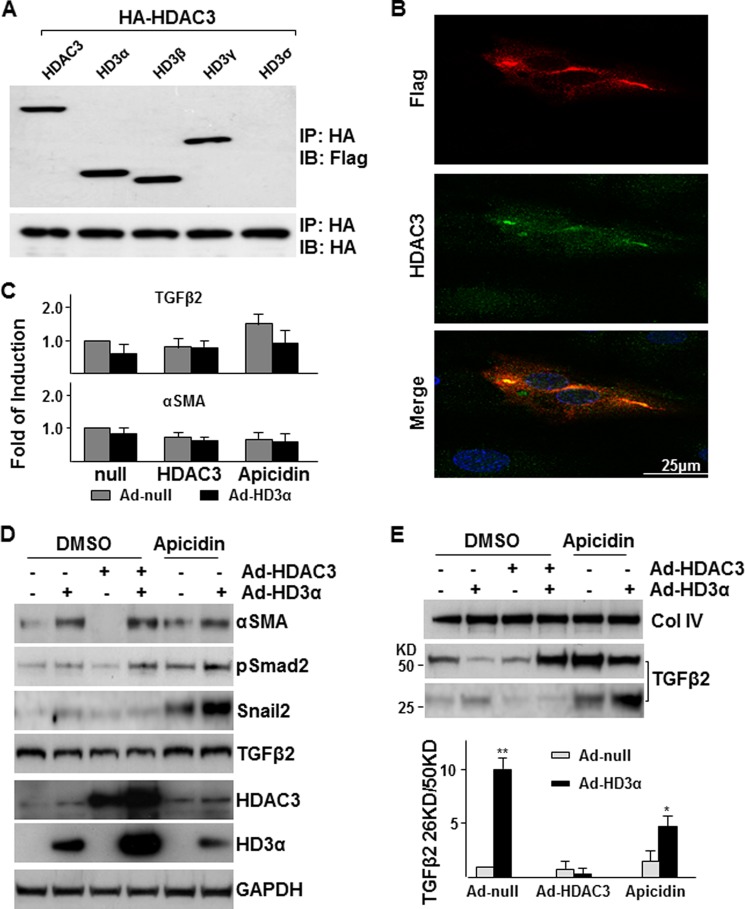
HD3α Modulates HDAC3 Functions
Being one of the isoforms, HD3α shares the same N terminus and part of the internal sequence of fully spliced HDAC3. Therefore, a natural question is whether HD3α can associate with and modulate HDAC3. Immunoprecipitation assay was first performed in 293 cells co-transfected with FLAG-tagged HDAC3 and all four isoform plasmids together with HA-tagged HDAC3 plasmid. As expected, HDAC3 could form a homodimer. HD3α, -β, and -γ isoforms, but not HD3δ, could form a heterodimer with HDAC3 (Fig. 6A). Immunofluorescence staining with antibodies against the different C-terminal domains (anti-HDAC3 (Sigma, H3034) against peptide from amino acids 411–428 of the HDAC3 protein and anti-HD3α raised against peptide derived from intron 12) confirmed the physical association of HDAC3 and HD3α, mainly in the cytoplasm (Fig. 6B). Interestingly, HDAC3 protein levels in HD3α-positive cells are much higher than that in HD3α-negative cells, suggesting that HD3α may stabilize HDAC3. To test whether there is a cross-talk or interaction between HDAC3 and HD3α on EndMT, overexpression of HDAC3 via adenoviral transfer and suppression of HDAC3 activity via selective inhibitor apicidin (40) were included in Ad-HD3α-induced EndMT assays. Quantitative RT-PCR assays revealed that neither overexpression of HDAC3 nor inhibition of HDAC3 activity had significant effect on TGFβ2 and αSMA mRNA levels (Fig. 6C). Overexpression of HDAC3 itself decreased the basal level of the EndMT phenotype as revealed by the decrease of αSMA, pSmad2, and Snail2 protein levels in cell lysate (Fig. 6D) and decreased TGFβ2 secretion/activation in the medium (Fig. 6E). In contrast, suppression of HDAC3 by the selective inhibitor apicidin induced TGFβ2 secretion/activation and the EndMT phenotype at basal levels and enhanced HD3α-induced EndMT (Fig. 6, D and E). A striking finding is that overexpression of HDAC3 enhances HD3α-induced TGFβ2 secretion, although TGFβ2 cleavage (Fig. 6E) and Smad2 phosphorylation and αSMA expression (Fig. 6D) were suppressed when HDAC3 and HD3α were expressed together. Overexpression of HD3α increased endogenous and exogenous HDAC3 protein level, and overexpression of HDAC3 could also increase exogenous HD3α protein level (Fig. 6D), confirming the immunofluorescence staining. These results suggest that HD3α may form a complex with HDAC3 and modulate HDAC3 function.
FIGURE 6.
HD3α modulates HDAC3 function. A, Western blot analysis detected the association of HDAC3 isoforms with HDAC3 in HEK293 cells. All isoforms except for HD3δ associated with HDAC3. B, HD3α physically associated with and stabilized HDAC3. HAECs were infected with Ad-HD3α at an MOI of 10 for 6 h in complete growth medium and cultured in serum-free medium for 24 h, followed by immunofluorescence staining with anti-HDAC3 (green) and anti-FLAG (for HD3α; red). C–E, effect of overexpression or suppression of HDAC3 on HD3α-induced EndMT. HAECs were co-infected with Ad-HD3α (10 MOI) and Ad-HDAC3 (10 MOI) or in the presence of 1 × 10−5 mol/liter apicidin in complete growth medium for 6 h and then cultured in M199 medium supplemented with 5 ng/ml insulin for 24 h, followed by quantitative RT-PCR analysis of TGFβ2 and αSMA mRNA levels (C), Western blot analysis of cell lysates (D), and Western blot analysis of conditioned media with the ratio of cleaved to precursor TGFβ2 indicated in the bottom panel (E). Ad-null is included as a control and to normalize variations in MOI. DMSO was included as a vehicle control. *, p < 0.05; **, p < 0.01. Data presented are representative of or the average of three independent experiments. IB, immunoblot; IP, immunoprecipitation.
DISCUSSION
EndMT is one type of EMT that refers only to vascular/cardiac endothelial cells undergoing mesenchymal transition, which contributes to cardiogenesis and a range of pathological fibrosis in human diseases (41–43). In the present study, we showed a novel finding that HDAC3 undergoes unconventional splicing to produce four different isoforms, of which HD3α contributes to stem cell differentiation and embryonic cardiovascular development. Overexpression of HD3α induces the EndMT phenotype in mature ECs through TGFβ2 activation. These findings provide new insights into the mechanism of EndMT.
Unconventional splicing is a common event occurring in mammalian cells in different cellular processes, which makes it possible for a single gene to produce different proteins with different or even opposite functions. In this study, the occurrence of GFP in HD3α/β-GFP-transfected cells, the specific band detected by Western blot, and specific staining in mouse embryos with HD3α/β antibody have demonstrated that intron 12 is kept as an additional exon. In addition to intron 12 retention, there seems to be recombination between exons because the spliced different exons cannot be produced from the routine splicing mechanisms. Due to the sequence differences in intron 12 among difference species, the HD3α isoform seems unique to Mus musculus. We have detected several isoforms from human HDAC3,4 which are different from those HDAC3 isoforms in M. musculus. Detailed investigations are still under way to decipher how intron 12 is kept as an additional exon, how the recombination events occur, and which human isoform possesses a function similar to that of HD3α.
HDAC3 forms a complex with co-repressors like N-CoR/SMRT and Ski, in which the co-repressors can activate HDAC3 deacetylase activity (44–46). These complexes may also contain other HDACs and suppress gene transcription via histone deacetylation. Structural analyses of HDAC3 protein indicate that the C-terminal domain is the deacetylase catalytic domain and is responsible for gene transcriptional repression, whereas the N-terminal domain is responsible for oligomerization of HDAC3 itself and interaction with other HDACs and proteins, such as Akt1 (25, 47). In addition to its deacetylase activity, HDAC3 may function as a scaffold in complexes, in which the N-terminal domain plays an essential role (48, 49). The HD3α isoform retains the N-terminal domain. Therefore, it can form a complex with HDAC3 protein. Indeed, we have detected the direct association of HD3α and HDAC3. The formation of the complex may modulate HDAC3 functions in several ways. First, HD3α may participate in an HDAC3 complex. Because the intron 12-derived C-terminal domain of HD3α is different from that of HDAC3, the involvement of HD3α in the complex may interfere with HDAC3 deacetylase activity, therefore attenuating the transcriptional repression effects. Second, HD3α may compete with HDAC3 for binding to associated proteins, excluding HDAC3 from some complexes and ablating both the catalytic and scaffolding roles of HDAC3. It has been reported that HDAC3 can associate with Akt and Smads (25, 45). In our study, overexpression of HDAC3 decreases Smad2 phosphorylation, suggesting a suppressive role of HDAC3 in TGFβ signaling. As expected, we found the association of HD3α with Akt1 and Smad3. In contrast to the suppressive effect of HDAC3 on Smad2 phosphorylation, HD3α increases Smad2 phosphorylation, suggesting that HD3α may interfere with HDAC3 interaction with Smad2 or modify the HDAC3-Smad2 complex. Further evidence for the competition hypothesis derives from the observation that overexpression of HD3α stabilizes HDAC3 protein. Immunofluorescence staining revealed that almost all HD3α proteins co-localize with HDAC3 and that the protein level of HDAC3 in HD3α-positive cells is much higher than that in HD3α-negative cells. When these two proteins were overexpressed together, the level of each protein was much higher than that in a single overexpression system. The formation of HD3α-HDAC3 complex may prevent the association with degrading enzymes, therefore increasing the stabilization. The dimerization of HD3α and HDAC3 may also introduce HDAC3 into new signal pathways via the interaction of the HD3α C-terminal domain with other proteins. A detailed investigation of the relationship between HD3α and HDAC3 is required.
Multiple factors have been reported to induce EndMT via several interactive signal pathways. The common factor is the TGFβ superfamily, in which TGFβ2 plays an indispensable role in EndMT in vivo (34). TGFβs bind to the TGFβ type II receptor, which recruits and phosphorylates the type I (ALK5) receptor. The latter activates Smad signaling pathways, leading to EndMT. In this study, we demonstrate that HD3α induces EndMT through TGFβ2. HD3α activates TGFβ2 in the extracellular matrix. The active TGFβ2 functions as an autocrine and paracrine factor to activate ALK5 receptor and Smad2/3 phosphorylation, leading to Snail2 up-regulation and EndMT. The evidence comes from the following observations. HD3α up-regulates Smad2/3 phosphorylation and Snail2 and mesenchymal marker expression, which can be abolished by suppression of the ALK5 receptor via SB435142. Higher amounts of cleaved TGFβ2 have been detected in conditioned medium from Ad-HD3α-infected HAECs as compared with Ad-null infected cells. Utilization of TGFβ2 neutralization antibody attenuates the HD3α-induced Smad2 phosphorylation and αSMA expression. Highly expressed αSMA was detected not only in HD3α-positive cells but also in adjacent cells. High levels of active TGFβ2 were detected when SB435142 was present. SB431542 binds to ALK5, blocking downstream events. The binding of SB435142 to ALK5 may inhibit the binding of TGFβ2 to the type II receptor in a negative feedback mechanism, therefore accumulating TGFβ2 in the medium.
HD3α-induced activation of TGFβ2 occurs in the extracellular matrix via protease-mediated cleavage. TGFβs are secreted in a latent form, associating with latent TGFβ-binding protein (50). Upon proteolytic cleavage, the latent TGFβ-binding protein and the latency-associated peptide are removed from TGFβ molecules, giving rise to 13-kDa singlet or 25-kDa dimer mature forms. Upon the overexpression of HD3α, no elevated TGFβs at mRNA and protein levels were discovered. However, the higher level of mature TGFβ2 was detected in Ad-HD3α-infected cell culture medium, suggesting that HD3α may activate certain proteases in the extracellular environment, whose expression and secretion were increased. Although TGFβ1 and TGFβ2 belong to the same family, their secretion mechanisms are different. TGFβ1 is secreted via exosomes, whereas TGFβ2 seems not to be. Therefore, the extracellular location or binding partners will be different for TGFβ1 from those of TGFβ2, and so is the activation mechanism involved (i.e. different proteases are responsible for the cleavage of TGFβ1 and TGFβ2). In this study, HD3α activates a few proteases, which locate in the cell surface or extracellular matrix but not in exosomes; therefore, they specifically initiate TGFβ2 but not TGFβ1 cleavage.
It has been reported that Smad-independent pathways are also involved in EndMT, such as MEK, PI3K/Akt, p38 and JNK MAPKs, and G-proteins (Rho). In this study, we also discovered that the PI3K/Akt pathway is essential for HD3α-induced EndMT. However, it may not activate TGFβ2, because genetic knockdown of PI3K or Akt1 ablated HD3α-induced αSMA expression but did not affect TGFβ2 activation. In p85 siRNA- and Akt1 siRNA-transfected cells, HD3α protein levels were significantly decreased, especially in the latter. Akt1 binds to HD3α in the absence or presence of LY294002, indicating that Akt1 phosphorylation is not involved in the binding. The complex formation between Akt1 and HD3α may contribute to the stabilization of HD3α protein. Considering that LY294002 abolishes TGFβ2 activation, Smad phosphorylation, and EndMT, whereas p85 or Akt1 knockdown does not affect TGFβ2 activation, it appears that other signal pathways may be involved, which are susceptible to LY294002.
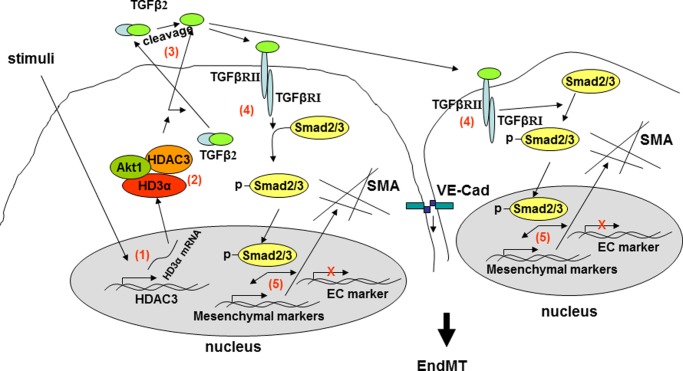
In summary, HDAC3 mRNA undergoes unconventional splicing, in which intron 12 is kept as an additional exon and recombination occurs between exons 3 and 9, giving rise to the HD3α isoform. The HD3α forms a complex with HDAC3 and Akt1 to promote TGFβ2 secretion and activation through a mechanism yet to be elucidated. The active TGFβ2 functions in an autocrine and paracrine manner to activate the ALK5-Smad2/3-Snail2 pathways, leading to EndMT (Fig. 7). This study provides the first evidence that HDAC3 mRNA can undergo unconventional splicing to modulate HDAC3 and induce EndMT. Further detailed investigation on the mechanisms involved in HDAC3 splicing and how the isoforms exert their actions will undoubtedly contribute to further insights into the role of HDAC3 in the maintenance of endothelium integrity.
FIGURE 7.
Schematic illustration of HDAC3 splicing-induced EndMT. Upon some kinds of stimuli, HDAC3 mRNA undergoes unconventional splicing (1), in which intron 12 is kept as an additional exon and recombination occurs between exon 3 and exon 9, giving rise to the HD3α isoform. The HD3α forms a complex with HDAC3 and Akt1 (2), which in turn promote TGFβ2 secretion and activation through unknown mechanisms (3). The active TGFβ2 functions in an autocrine and paracrine manner to activate Alk5-Smad2/3-Snail2 pathways (4 and 5), leading to EndMT.
This work was supported by grants from the British Heart Foundation and the Oak Foundation and National Natural Science Foundation of China Grant 81300116.
L. Zeng, G. Wang, D. Ummarino, A. Margariti, Q. Xu, Q. Xiao, W. Wang, Z. Zhang, X. Yin, M. Mayr, G. Cockerill, J. Y. Li, S. Chien, Y. Hu, and Q. Xu, unpublished data.
- EC
- endothelial cell
- EndMT
- endothelial-to-mesenchymal transition
- αSMA
- α-smooth muscle actin
- VE-Cad
- VE-cadherin
- dpc
- days postcoitus
- HDAC
- histone deacetylase
- ES
- embryonic stem
- HAEC
- human aortic endothelial cell
- DM
- differentiation medium
- MOI
- multiplicity of infection
- DES
- differentiated embryonic stem
- Ad
- adenovirus.
REFERENCES
- 1. Chi J. T., Chang H. Y., Haraldsen G., Jahnsen F. L., Troyanskaya O. G., Chang D. S., Wang Z., Rockson S. G., van de Rijn M., Botstein D., Brown P. O. (2003) Endothelial cell diversity revealed by global expression profiling. Proc. Natl. Acad. Sci. U.S.A. 100, 10623–10628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Markwald R. R., Fitzharris T. P., Smith W. N. (1975) Structural analysis of endocardial cytodifferentiation. Dev. Biol. 42, 160–180 [DOI] [PubMed] [Google Scholar]
- 3. Markwald R. R., Fitzharris T. P., Manasek F. J. (1977) Structural development of endocardial cushions. Am. J. Anat. 148, 85–119 [DOI] [PubMed] [Google Scholar]
- 4. Thiery J. P., Sleeman J. P. (2006) Complex networks orchestrate epithelial-mesenchymal transitions. Nat. Rev. Mol. Cell Biol. 7, 131–142 [DOI] [PubMed] [Google Scholar]
- 5. Nakajima Y., Yamagishi T., Hokari S., Nakamura H. (2000) Mechanisms involved in valvuloseptal endocardial cushion formation in early cardiogenesis. Roles of transforming growth factor (TGF)-β and bone morphogenetic protein (BMP). Anat. Rec. 258, 119–127 [DOI] [PubMed] [Google Scholar]
- 6. Wang J., Sridurongrit S., Dudas M., Thomas P., Nagy A., Schneider M. D., Epstein J. A., Kaartinen V. (2005) Atrioventricular cushion transformation is mediated by ALK2 in the developing mouse heart. Dev. Biol. 286, 299–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Armulik A., Abramsson A., Betsholtz C. (2005) Endothelial/pericyte interactions. Circ. Res. 97, 512–523 [DOI] [PubMed] [Google Scholar]
- 8. Zeisberg E. M., Tarnavski O., Zeisberg M., Dorfman A. L., McMullen J. R., Gustafsson E., Chandraker A., Yuan X., Pu W. T., Roberts A. B., Neilson E. G., Sayegh M. H., Izumo S., Kalluri R. (2007) Endothelial-to-mesenchymal transition contributes to cardiac fibrosis. Nat. Med. 13, 952–961 [DOI] [PubMed] [Google Scholar]
- 9. Zeisberg E. M., Potenta S. E., Sugimoto H., Zeisberg M., Kalluri R. (2008) Fibroblasts in kidney fibrosis emerge via endothelial-to-mesenchymal transition. J. Am. Soc. Nephrol. 19, 2282–2287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Arciniegas E., Frid M. G., Douglas I. S., Stenmark K. R. (2007) Perspectives on endothelial-to-mesenchymal transition. Potential contribution to vascular remodeling in chronic pulmonary hypertension. Am. J. Physiol. Lung Cell Mol. Physiol. 293, L1–L8 [DOI] [PubMed] [Google Scholar]
- 11. Potenta S., Zeisberg E., Kalluri R. (2008) The role of endothelial-to-mesenchymal transition in cancer progression. Br. J. Cancer 99, 1375–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zeisberg E. M., Potenta S., Xie L., Zeisberg M., Kalluri R. (2007) Discovery of endothelial to mesenchymal transition as a source for carcinoma-associated fibroblasts. Cancer Res. 67, 10123–10128 [DOI] [PubMed] [Google Scholar]
- 13. Potts J. D., Runyan R. B. (1989) Epithelial-mesenchymal cell transformation in the embryonic heart can be mediated, in part, by transforming growth factor β. Dev. Biol. 134, 392–401 [DOI] [PubMed] [Google Scholar]
- 14. Armstrong E. J., Bischoff J. (2004) Heart valve development. Endothelial cell signaling and differentiation. Circ. Res. 95, 459–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Meadows K. N., Iyer S., Stevens M. V., Wang D., Shechter S., Perruzzi C., Camenisch T. D., Benjamin L. E. (2009) Akt promotes endocardial-mesenchyme transition. J. Angiogenes. Res. 1, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen P. Y., Qin L., Barnes C., Charisse K., Yi T., Zhang X., Ali R., Medina P. P., Yu J., Slack F. J., Anderson D. G., Kotelianski V., Wang F., Tellides G., Simons M. (2012) FGF regulates TGF-β signaling and endothelial-to-mesenchymal transition via control of let-7 miRNA expression. Cell Rep. 2, 1684–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. de Ruijter A. J., van Gennip A. H., Caron H. N., Kemp S., van Kuilenburg A. B. (2003) Histone deacetylases (HDACs). Characterization of the classical HDAC family. Biochem. J. 370, 737–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gallinari P., Di Marco S., Jones P., Pallaoro M., Steinkühler C. (2007) HDACs, histone deacetylation, and gene transcription. From molecular biology to cancer therapeutics. Cell Res. 17, 195–211 [DOI] [PubMed] [Google Scholar]
- 19. Kekatpure V. D., Dannenberg A. J., Subbaramaiah K. (2009) HDAC6 modulates Hsp90 chaperone activity and regulates activation of aryl hydrocarbon receptor signaling. J. Biol. Chem. 284, 7436–7445 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20. Zilberman Y., Ballestrem C., Carramusa L., Mazitschek R., Khochbin S., Bershadsky A. (2009) Regulation of microtubule dynamics by inhibition of the tubulin deacetylase HDAC6. J. Cell Sci. 122, 3531–3541 [DOI] [PubMed] [Google Scholar]
- 21. Margariti A., Xiao Q., Zampetaki A., Zhang Z., Li H., Martin D., Hu Y., Zeng L., Xu Q. (2009) Splicing of HDAC7 modulates the SRF-myocardin complex during stem-cell differentiation towards smooth muscle cells. J. Cell Sci. 122, 460–470 [DOI] [PubMed] [Google Scholar]
- 22. Margariti A., Zampetaki A., Xiao Q., Zhou B., Karamariti E., Martin D., Yin X., Mayr M., Li H., Zhang Z., De Falco E., Hu Y., Cockerill G., Xu Q., Zeng L. (2010) Histone deacetylase 7 controls endothelial cell growth through modulation of β-catenin. Circ. Res. 106, 1202–1211 [DOI] [PubMed] [Google Scholar]
- 23. Emiliani S., Fischle W., Van Lint C., Al-Abed Y., Verdin E. (1998) Characterization of a human RPD3 ortholog, HDAC3. Proc. Natl. Acad. Sci. U.S.A. 95, 2795–2800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bhaskara S., Chyla B. J., Amann J. M., Knutson S. K., Cortez D., Sun Z. W., Hiebert S. W. (2008) Deletion of histone deacetylase 3 reveals critical roles in S phase progression and DNA damage control. Mol. Cell 30, 61–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zampetaki A., Zeng L., Margariti A., Xiao Q., Li H., Zhang Z., Pepe A. E., Wang G., Habi O., deFalco E., Cockerill G., Mason J. C., Hu Y., Xu Q. (2010) Histone deacetylase 3 is critical in endothelial survival and atherosclerosis development in response to disturbed flow. Circulation 121, 132–142 [DOI] [PubMed] [Google Scholar]
- 26. Zeng L., Xiao Q., Margariti A., Zhang Z., Zampetaki A., Patel S., Capogrossi M. C., Hu Y., Xu Q. (2006) HDAC3 is crucial in shear- and VEGF-induced stem cell differentiation toward endothelial cells. J. Cell Biol. 174, 1059–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xiao Q., Zeng L., Zhang Z., Margariti A., Ali Z. A., Channon K. M., Xu Q., Hu Y. (2006) Sca-1+ progenitors derived from embryonic stem cells differentiate into endothelial cells capable of vascular repair after arterial injury. Arterioscler. Thromb. Vasc. Biol. 26, 2244–2251 [DOI] [PubMed] [Google Scholar]
- 28. Zeng L., Zampetaki A., Margariti A., Pepe A. E., Alam S., Martin D., Xiao Q., Wang W., Jin Z. G., Cockerill G., Mori K., Li Y. S., Hu Y., Chien S., Xu Q. (2009) Sustained activation of XBP1 splicing leads to endothelial apoptosis and atherosclerosis development in response to disturbed flow. Proc. Natl. Acad. Sci. U.S.A. 106, 8326–8331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kumarswamy R., Volkmann I., Jazbutyte V., Dangwal S., Park D. H., Thum T. (2012) Transforming growth factor-β-induced endothelial-to-mesenchymal transition is partly mediated by microRNA-21. Arterioscler. Thromb. Vasc. Biol. 32, 361–369 [DOI] [PubMed] [Google Scholar]
- 30. Mihira H., Suzuki H. I., Akatsu Y., Yoshimatsu Y., Igarashi T., Miyazono K., Watabe T. (2012) TGF-β-induced mesenchymal transition of MS-1 endothelial cells requires Smad-dependent cooperative activation of Rho signals and MRTF-A. J. Biochem. 151, 145–156 [DOI] [PubMed] [Google Scholar]
- 31. Medici D., Potenta S., Kalluri R. (2011) Transforming growth factor-β2 promotes Snail-mediated endothelial-mesenchymal transition through convergence of Smad-dependent and Smad-independent signalling. Biochem. J. 437, 515–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang J., Kuiatse I., Lee A. V., Pan J., Giuliano A., Cui X. (2010) Sustained c-Jun-NH2-kinase activity promotes epithelial-mesenchymal transition, invasion, and survival of breast cancer cells by regulating extracellular signal-regulated kinase activation. Mol. Cancer Res. 8, 266–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Derynck R., Zhang Y. E. (2003) Smad-dependent and Smad-independent pathways in TGF-β family signalling. Nature 425, 577–584 [DOI] [PubMed] [Google Scholar]
- 34. Azhar M., Runyan R. B., Gard C., Sanford L. P., Miller M. L., Andringa A., Pawlowski S., Rajan S., Doetschman T. (2009) Ligand-specific function of transforming growth factor beta in epithelial-mesenchymal transition in heart development. Dev. Dyn. 238, 431–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schniewind B., Groth S., Sebens Muerkoster S., Sipos B., Schäfer H., Kalthoff H., Fändrich F., Ungefroren H. (2007) Dissecting the role of TGF-β type I receptor/ALK5 in pancreatic ductal adenocarcinoma. Smad activation is crucial for both the tumor suppressive and prometastatic function. Oncogene 26, 4850–4862 [DOI] [PubMed] [Google Scholar]
- 36. Clayton A., Mitchell J. P., Court J., Mason M. D., Tabi Z. (2007) Human tumor-derived exosomes selectively impair lymphocyte responses to interleukin-2. Cancer Res. 67, 7458–7466 [DOI] [PubMed] [Google Scholar]
- 37. Théry C., Regnault A., Garin J., Wolfers J., Zitvogel L., Ricciardi-Castagnoli P., Raposo G., Amigorena S. (1999) Molecular characterization of dendritic cell-derived exosomes. Selective accumulation of the heat shock protein hsc73. J. Cell Biol. 147, 599–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Larue L., Bellacosa A. (2005) Epithelial-mesenchymal transition in development and cancer. Role of phosphatidylinositol 3′-kinase/AKT pathways. Oncogene 24, 7443–7454 [DOI] [PubMed] [Google Scholar]
- 39. Hatipoglu O. F., Hirohata S., Cilek M. Z., Ogawa H., Miyoshi T., Obika M., Demircan K., Shinohata R., Kusachi S., Ninomiya Y. (2009) ADAMTS1 is a unique hypoxic early response gene expressed by endothelial cells. J. Biol. Chem. 284, 16325–16333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Strobl J. S., Cassell M., Mitchell S. M., Reilly C. M., Lindsay D. S. (2007) Scriptaid and suberoylanilide hydroxamic acid are histone deacetylase inhibitors with potent anti-Toxoplasma gondii activity in vitro. J. Parasitol. 93, 694–700 [DOI] [PubMed] [Google Scholar]
- 41. Piera-Velazquez S., Li Z., Jimenez S. A. (2011) Role of endothelial-mesenchymal transition (EndoMT) in the pathogenesis of fibrotic disorders. Am. J. Pathol. 179, 1074–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chang A. C., Fu Y., Garside V. C., Niessen K., Chang L., Fuller M., Setiadi A., Smrz J., Kyle A., Minchinton A., Marra M., Hoodless P. A., Karsan A. (2011) Notch initiates the endothelial-to-mesenchymal transition in the atrioventricular canal through autocrine activation of soluble guanylyl cyclase. Dev. Cell 21, 288–300 [DOI] [PubMed] [Google Scholar]
- 43. Aisagbonhi O., Rai M., Ryzhov S., Atria N., Feoktistov I., Hatzopoulos A. K. (2011) Experimental myocardial infarction triggers canonical Wnt signaling and endothelial-to-mesenchymal transition. Dis. Model Mech. 4, 469–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Li J., Wang J., Wang J., Nawaz Z., Liu J. M., Qin J., Wong J. (2000) Both corepressor proteins SMRT and N-CoR exist in large protein complexes containing HDAC3. EMBO J. 19, 4342–4350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tabata T., Kokura K., Ten Dijke P., Ishii S. (2009) Ski co-repressor complexes maintain the basal repressed state of the TGF-β target gene, SMAD7, via HDAC3 and PRMT5. Genes Cells 14, 17–28 [DOI] [PubMed] [Google Scholar]
- 46. Guenther M. G., Barak O., Lazar M. A. (2001) The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol. Cell Biol. 21, 6091–6101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yang W. M., Tsai S. C., Wen Y. D., Fejer G., Seto E. (2002) Functional domains of histone deacetylase-3. J. Biol. Chem. 277, 9447–9454 [DOI] [PubMed] [Google Scholar]
- 48. Alam S., Li H., Margariti A., Martin D., Zampetaki A., Habi O., Cockerill G., Hu Y., Xu Q., Zeng L. (2011) Galectin-9 protein expression in endothelial cells is positively regulated by histone deacetylase 3. J. Biol. Chem. 286, 44211–44217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Togi S., Kamitani S., Kawakami S., Ikeda O., Muromoto R., Nanbo A., Matsuda T. (2009) HDAC3 influences phosphorylation of STAT3 at serine 727 by interacting with PP2A. Biochem. Biophys. Res. Commun. 379, 616–620 [DOI] [PubMed] [Google Scholar]
- 50. Miyazono K., Olofsson A., Colosetti P., Heldin C. H. (1991) A role of the latent TGF-β1-binding protein in the assembly and secretion of TGF-β1. EMBO J. 10, 1091–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]