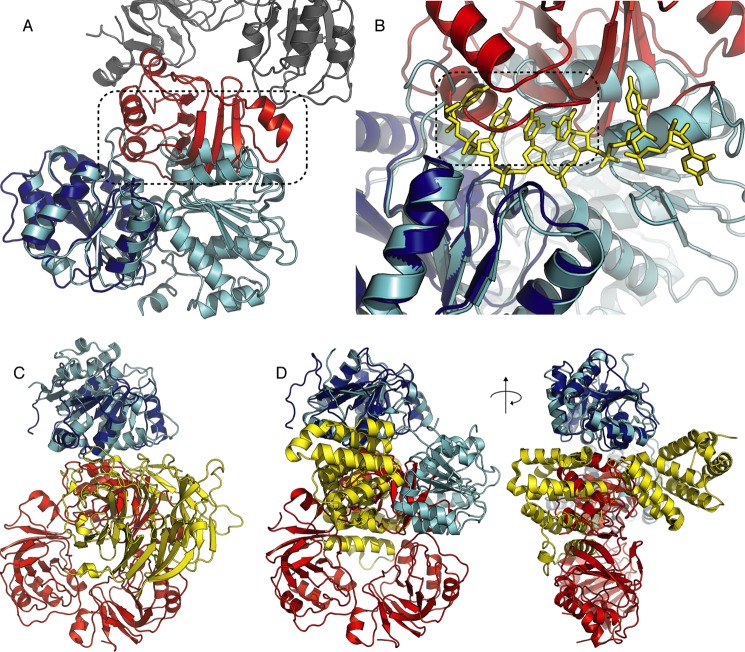
FIGURE 6.
Comparison of the RraA/SrmB structure and other DEAD-box protein complexes. A, B RraA bound to SrmB would prevent RecA-like domain closure due to steric clash (highlighted by dashed frame). SrmB is colored blue, the RraA protomer interacting with SrmB is shown in red and the other RraA protomers are shown in gray. The Dbp5 structure in closed conformation (PDB code 3FHT), superimposed onto SrmB CTD is shown in cyan with RNA colored yellow and shown in stick representation. The root mean square deviation for the fit is 1.34 Å. B, RraA binding may impede RNA binding by SrmB. The RraA loop consisting of residues 53–59 would occlude the predicted RNA binding surface of SrmB (highlighted by dashed frame). C, comparison of the RraA-SrmB and Dbp5-NUP214 complexes. The reference frame for the overlay is the SrmB CTD (blue) and the Dbp5 NTD (cyan). RraA is red and NUP214 is yellow (PDB code 3FHC). D, comparison of the RraA-SrmB and eIF4A-PDCD4 complexes. Superposition of SrmB CTD (blue) on eIF4A NTD (cyan). PDCD4 is shown in yellow (PDB code 2ZU6). The overlays are shown from two viewpoints related by a 90° rotation.