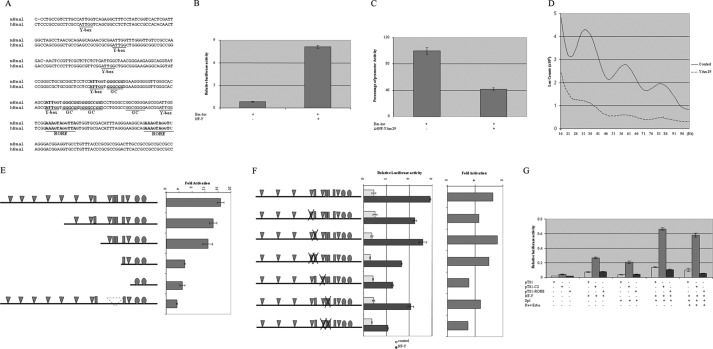
FIGURE 2.
NF-Y is the functional transcription activator of Bmal1 gene. A, alignment of human (hBmal) and mouse (mBmal) Bmal1 promoter proximal region. The important and highly conserved transcription factor-binding sites are shown in boldface and underlined. B, Bmal1 luciferase (Bm-luc) reporter gene (d318) was co-transfected with NF-Y or empty pSG5 vectors into NIH 3T3 cells. Luciferase activity was measured 48 h later. C, Bmal1 luciferase reporter gene (d318) was co-transfected with dominant negative NF-YA or empty pSG5 vectors into NIH 3T3 cells. Luciferase activity was measured 48 h later. The remaining luciferase activity was shown. D, NIH 3T3 cells were transfected with Bmal1 promoter construct in combination with control vector or pSG5-YAm29 and treated the same way as in Fig. 1. E, 5′-truncated Bmal1 promoter constructs were co-transfected with NF-Y or empty pSG5 vector. The ratio of luciferase activity of each reporter with and without NF-Y was shown. F, Bmal1 promoter (d318) reporter constructs with mutated different CCAAT-box or/and Sp1 site were co-transfected with either pSG5 or NF-Y expression constructs. The relative luciferase activity (left panel) and -fold of activation by NF-Y (right panel) were shown. G, pT81-carrying the proximal CCAAT/GC-box/GC-box (C2) and RORα-binding sites of mouse Bmal1 promoter were co-transfected with NF-Y, Sp1, and/or Rev-Erbα. The luciferase activity relative to β-galactosidase activity was shown. Error bars in B, C, E, F, and G indicate mean ± S.E.