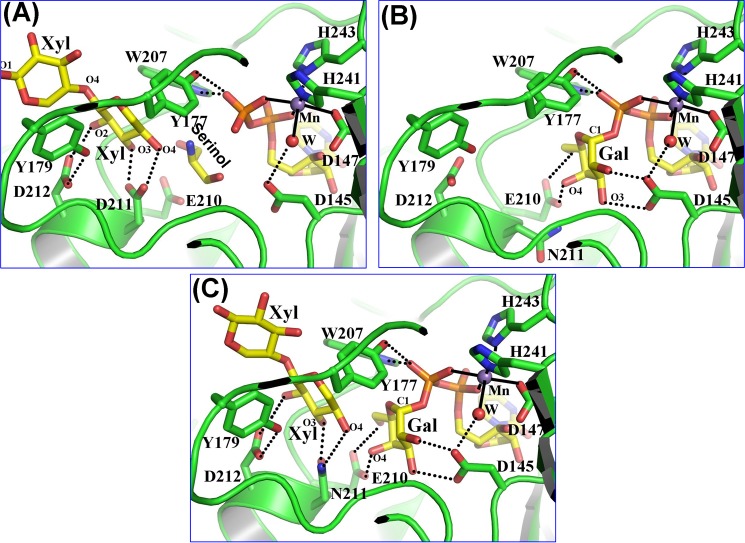
FIGURE 2.
Substrate binding to the Drosophila β4GalT7 molecule. The protein molecule is represented by the green graphic, with the interacting residues shown as colored sticks. The substrate molecules UDP-Gal and xylobiose are shown as yellow sticks. The manganese and water molecules are shown as magenta and red spheres. The hydrogen bonds are shown in black dotted lines, and the manganese coordination bond is shown in solid black lines. A, binding of a xylobiose molecule to the Drosophila β4GalT7 molecule in the closed conformation. The acceptor substrate forms four hydrogen bonds with the protein molecule. The C5 and O5 atoms of the acceptor xylose molecule form strong hydrophobic interactions with the aromatic side chain of the Tyr177 residue. Thus, the presence of this residue determines the enzyme acceptor specificity for xylose instead of glucose. The extended xylose moiety forms stacking interactions with the aromatic side chain of the Tyr179 residue. When the PG acceptor substrate binds to the enzyme, with its O-linked β-xylose moiety bound in the acceptor substrate binding site, this extended binding site is likely to facilitate the binding of the protein moiety of the PG acceptor molecule. B, UDP-gal binding to the D211N β4GalT7 mutant protein. The binding of the UDP moiety in the present crystal structure and in the human β4GalT7 closed conformation crystal structure (Fig. 1B) is quite similar (supplemental Fig. S3). The Gal moiety of the bound UDP-Gal forms seven hydrogen bonds with the protein molecule. These hydrogen-bonding interactions are similar to those that occur when UDP-Gal binds to the β4GalT1 molecule. C, the binding of the xylobiose together with UDP-Gal to Drosophila D211N β4GalT7 molecule. The molecular interactions of the substrates with the protein molecule are similar to their individual binding to the enzyme (panels A and B). In the present structure, the O4 oxygen atom of the xylobiose molecule forms a hydrogen bond with the Nϵ2 amino group of the Asn211 residue, which is in contrast to the panel A where the same O4 oxygen atom forms a hydrogen bond with the Oϵ2 atom of the Asp211 residue. The binding of the xylobiose does not seem to affect the binding of the UDP-Gal molecule or vice versa.