Figure 3.
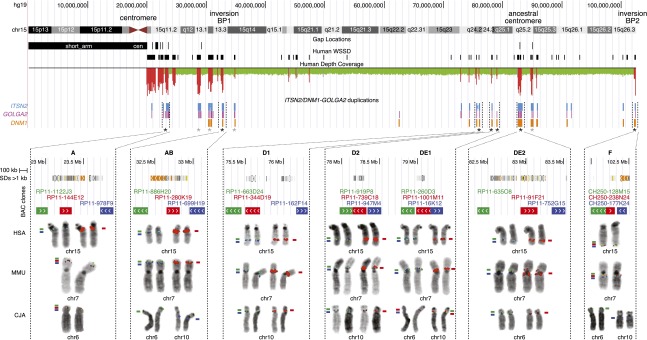
Relationships between the ITSN2/DNM1–GOLGA2 duplications and chromosome 15 evolutionary hotspots. Gap locations, WSSD-positive regions, depth-of-coverage data, and locations (BLAT search; Kent 2002) of the ITSN2, DMN1, and GOLGA2 duplications are shown for the sequence of chromosome 15 (hg19/build37). (Note, only a few LCR15 copies lack one of the three components.) Asterisks indicate the 11 loci analyzed through FISH. The BAC triplets were chosen as follows: one BAC (in red) spanning LCR15 and the other two at the single-copy regions immediately upstream (in green) and downstream (in blue) of the duplication cluster. SDs (hg19) and FISH mapping data on human, macaque, and marmoset chromosomes are shown for seven loci (named A, AB, D1, D2, DE1, DE2, and F, and indicated by a black asterisk). For BAC clones spanning LCR15 sequences (red-colored) only signals co-mapping with the single-copy clones (green and blue colored) are shown. BAC clones designed in single-copy regions of loci AB and F co-hybridized in human and macaque, whereas in marmoset they hybridized on chromosomes 6 and 10, respectively, for green- and blue-colored clones; no signal is on marmoset chromosomes for the red-colored BAC. Locus DE1 single-copy BAC clones mapped to marmoset chromosomes 10 and 6, respectively; the red-colored BAC clone showed signal on both chromosomes. Locus DE2 single-copy BAC clones hybridized to the macaque 7p and 7q pericentromeric regions, respectively.
