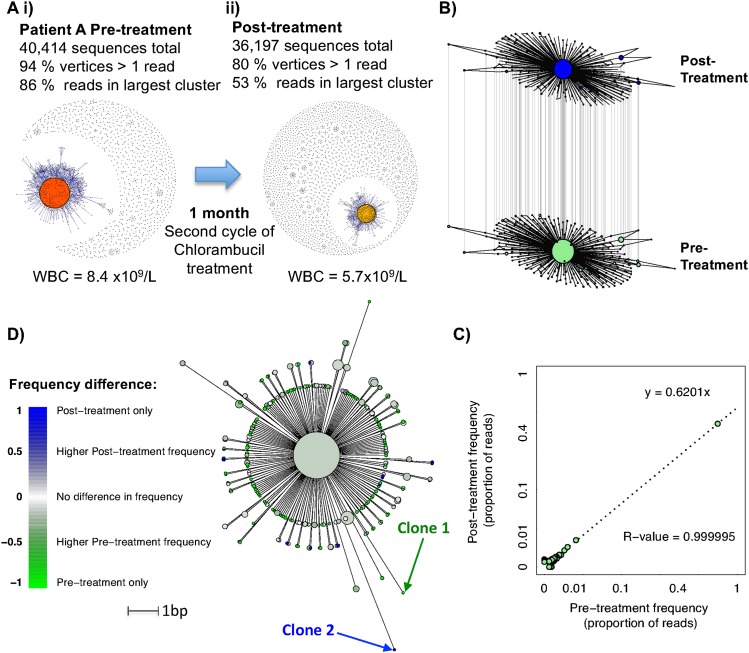
Figure 4.
B-cell leukemic clonal evolution. (A) The B-cell sequence networks for patient A with chronic lymphocytic leukemia for samples (i) prior to and (ii) after second cycle of Chlorambucil treatment, separated by 1 mo with corresponding white blood cell counts. (B) All sequences from the dominant clusters from both temporal samples were used to generate a composite network, and the differential frequencies at each time point are indicated by the relative vertex sizes. (C) Correlation between the proportional frequencies of each unique BCR within the dominant clones of patient A with corresponding R-value and linear regression equation. (D) An unrooted maximum parsimony tree was generated showing the relationships between sequences that were observed at least six times between the pre- and post-treatment samples, where the branch lengths are proportional to the number of varying bases (evolutionary distance). The tip colors show the relative difference in sequence abundance between the different time points, where green indicates observation of sequence primarily in the pretreatment sample, blue indicates predominant observations in the post-treatment sample, and white indicates no change in frequency. Clones 1 and 2 refer to examples of BCRs observed only in the pre- or post-treatment samples, respectively.