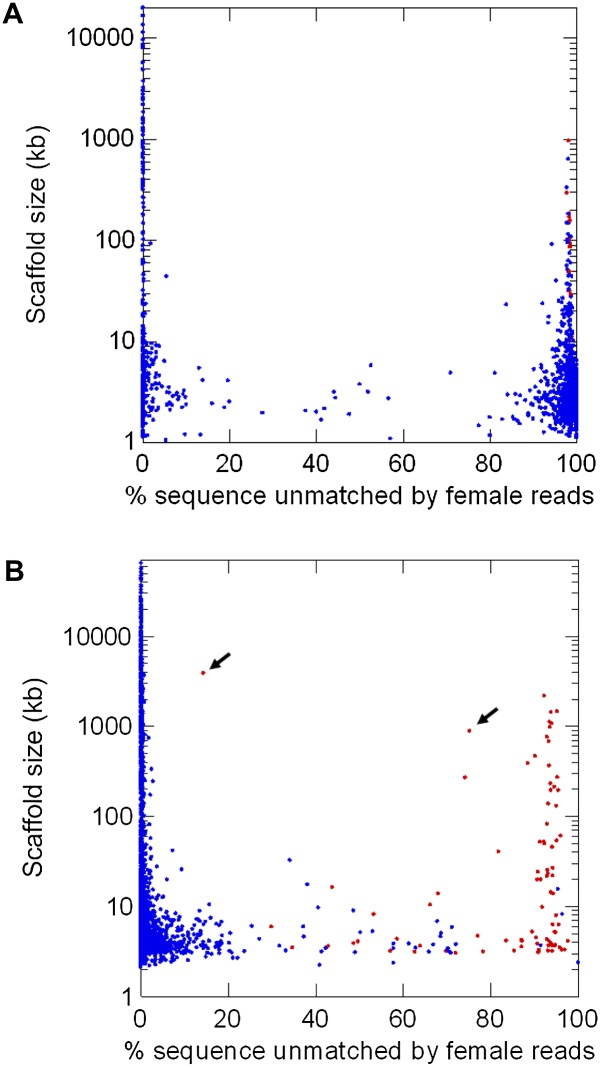
Figure 3.
Identification of Y-linked scaffolds in the D. virilis (A) and human (B) genomes (scatter plot). The figure complements Figure 2A,B, adding the scaffold size and mapping. Each dot represents one scaffold in the assembled genomes; red dots are scaffolds previously known to be Y-linked, and blue dots are unmapped or not Y-linked scaffolds. (Abscissa) proportion of scaffold sequence not matched by female short reads (in percentage unmatched single-copy k-mers); (ordinate) scaffold size. (A) D. virilis genome (CA assembly).There are 15 intermediate scaffolds (defined as those having between 25% and 80% unmatched single-copy k-mers); they are all small (total size: 40.4 kb ; 0.02% of the genome), and they seem to originate from misassembled repeats (see Results). (B) Human genome (HuRef assembly). There are 51 intermediate scaffolds; all are Y-linked and many of them could be traced to misassembled segmental duplications involving the Y chromosome, such as the two marked with arrows (accessions DS486171 and DS486351; see Results).