Figure 2. Putative glycosomal pathway scheme.
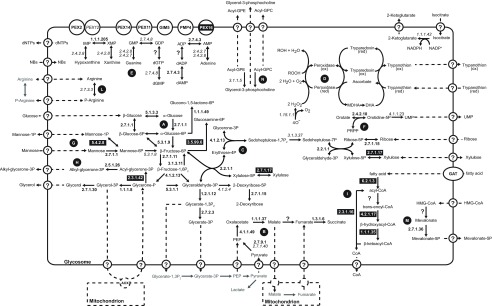
The scheme summarizes metabolic pathways identified so far in the glycosomes of Leishmania and T. brucei. EC numbers of enzymes identified only in the Leishmania glycosome are indicated in black boxes with white text, those identified only in T. brucei are indicated in italics, and those found in the glycosome of both species are indicated in bold. Predicted transport processes across the glycosomal membrane are indicated by the circled question marks and dashed arrows. Letters in black circles indicate the different metabolic pathways as follows: A: glycolysis; B: succinic fermentation; C: pentose phosphate pathway; D: superoxide and trypanothione metabolism; E: purine salvage; F: pyrimidine metabolism; G: mannose metabolism; H: glycerolipid biosynthesis; I: β-oxidation of fatty acids; L: phosphoarginine metabolism; M: mevalonate pathway; N: phospholipid degradation. Abbreviations used are: Acyl-GPC, 1-acyl-glycero-phosphocholine; Acyl-GPE, 1-acyl-glycero-phosphoethanolamine; AOX, alternative oxidase; DHA, dehydroascorbate; MDHA, monodehydroxyascorbate; NB, nucleobases; PRPP, 5-phosphoribosyl 1-pyrophosphate.
