Abstract
Objectives and design:
Developing an effective HIV-1 vaccine that elicits broadly neutralizing HIV-1 human antibodies (bnAbs) remains a challenging goal. Extensive studies on HIV-1 have revealed various strategies employed by the virus to escape host immune surveillance. Here, we investigated the human antibody gene repertoires of uninfected and HIV-1-infected individuals at genomic DNA (gDNA) and cDNA levels by deep sequencing followed by high-throughput sequence analysis to determine the frequencies of putative germline antibody genes of known HIV-1 monoclonal bnAbs (bnmAbs).
Methods:
Combinatorial gDNA and cDNA antibody libraries were constructed using the gDNAs and mRNAs isolated from uninfected and HIV-1-infected human peripheral blood mononuclear cells (PBMCs). All libraries were deep sequenced and sequences analysed using IMGT/HighV-QUEST software (http://imgt.org/HighV-QUEST/index.action). The frequencies of putative germline antibodies of known bnmAbs in the gDNA and cDNA libraries were determined.
Results and conclusion:
The human gDNA antibody libraries were more diverse in heavy and light chain V-gene lineage usage than the cDNA libraries, indicating that the human gDNA antibody gene repertoires may have more potential than the cDNA repertoires to develop HIV-1 bnAbs. The frequencies of the heavy and kappa and lambda light chain variable regions with identical V(D)J recombinations to known HIV-1 bnmAbs were extremely low in human antibody gene repertoires. However, we found relatively high frequencies of the heavy and kappa and lambda light chain variable regions that used the same V-genes and had the same CDR3 lengths as known HIV-1 bnmAbs regardless of (D)J-gene usage. B-cells bearing B-cell receptors of such heavy and kappa and lambda light chain variable regions may be stimulated to induce HIV-1 bnAbs.
Keywords: antibody somatic maturation, cDNA, genomic DNA, germline antibodies, HIV-1, neutralizing antibodies
Introduction
Since the discovery of HIV-1 in the early 1980s, an effective HIV-1 vaccine that can elicit bnAbs has yet to be developed. Extensive studies on HIV-1 have revealed various mechanisms for viral escape from human immune surveillance, including genetic alterations, oligomerization of envelope (Env) glycoproteins, heavy glycosylation and conformational masking [1–7]. But little is known about the potential of the human immune system to develop HIV-1 bnAbs. About 10–30% HIV-1 infected individuals develop cross-clade neutralizing Abs in natural infection, but only 1–3% individuals develop high titres of potent bnAbs after years of chronic infection. Enormous efforts have been made to isolate bnmAbs from HIV-1 infected ‘elite controllers’ whose sera exhibit high titres of broad neutralization activity. Four well known bnmAbs, b12, 2G12, 2F5 and 4E10, were identified more than a decade ago [8–11]. Many new and more potent bnmAbs were reported in the past 3 years, including PG9/16, HJ16, VRC01–03, VRC01-like Abs, PGTs and 10e8 [12–19]. Approximately 12 bnmAbs have been cocrystalized with Env and their neutralizing epitopes determined [18,20–26]. However, immunogens designed to include the neutralizing determinants of several HIV-1 bnmAbs have not been successful in inducing the same or similar bnAbs.
We and others have demonstrated that known HIV-1 bnmAbs had uncommon properties compared with bnmAbs against other microbes, including extensive somatic maturation and lack of measurable binding activity of their putative germline antibodies to Envs [13,15,16,18,27,28], suggesting that HIV-1 infection or vaccination with HIV-1 Envs may not initiate the somatic maturation processes of the putative germline Abs to bnAbs. Deep sequencing of the cDNA-PCR products of memory B cells obtained from several ‘elite controllers’ at different time points postinfection further revealed the limited use of heavy chain V-gene (IGHV) lineages for developing HIV-1 bnAbs in a single infected individual [17,18,29]. The infrequency of developing bnAbs in natural infection, the uncommon properties of known HIV-1 bnmAbs, the confined usage of IGHV lineages for developing bnAbs in a single infected individual and the failure in inducing the same or similar bnmAbs by vaccine immunogens prompted us to investigate the human antibody gene repertoire for the availability of the putative germline antibody genes of known HIV-1 bnmAbs at both genomic DNA (gDNA) and cDNA levels and the possibility for existence of alternative germline antibody genes that may potentially mature to HIV-1 bnAbs. The gDNAs of peripheral B cells in uninfected human individuals presumably possess the initial rearranged antibody gene repertoire for affinity maturation upon infection or vaccination. We hypothesized that antibody gene repertoire at the gDNA level may be more diverse than that at the cDNA level. Therefore, we developed a methodology for constructing large combinatorial gDNA antibody libraries and constructed one large nonimmune gDNA library using the PMBCs obtained from 300 uninfected healthy humans and three immune gDNA libraries using the PBMCs obtained from three HIV-1-infected ‘elite controllers’. Their corresponding cDNA libraries were simultaneously constructed. We compared the antibody gene repertoires of the four gDNA antibody libraries with those of the corresponding cDNA libraries by deep sequencing. Sequence analysis results suggest that the frequencies of the putative germline antibody genes of known HIV-1 bnmAbs were extremely low, but alternative germline antibody genes may be explored to elicit HIV-1 bnAbs.
Materials and methods
Preparation of human peripheral blood mononuclear cells
The PBMCs of healthy volunteers were obtained from The University of Hong Kong (Hong Kong, China). PBMCs from patient one (pt1) were obtained from Aaron Diamond AIDS Research Center, Rockefeller University (New York, USA). PBMCs from patients 2 and 3 (pt2 and pt3) were obtained from National Center for AIDS/STD Control and Prevention, China CDC (Beijing, China). All these experiments were approved by ethical committees of the respective institutes, and conducted according to local guidelines and regulations. All three patients were elite controllers. Pt1 was infected with clade B virus whose serum exhibited high titres of broadly neutralizing activity, and a panel of bnmAbs were isolated from pt1 PBMCs by single memory B-cell sorting using gp120 C-C core protein as a bait (pt1 in [18,28]). Pt2 and pt3 were infected with clade B’ virus and naive to antiretroviral therapy [30]. The sera of pt2 and pt3 also exhibited cross-clade neutralization activity (data not shown). Heparinized whole blood samples were used to isolate human PBMCs by Ficoll density gradient separation.
Isolation of genomic DNA, total RNA and mRNA from human peripheral blood mononuclear cells
Five to ten million PBMCs were collected from each 5 ml of heparinized whole blood and were used to isolate gDNA and total RNA using Allprep DNA/RNA Mini kits (Qiagen, Hilden, Germany) following the protocol provided by the manufacturer. The gDNA was used as a template for amplification of heavy chain variable regions and kappa and lambda light chain variable regions by semi-nested PCR for construction of gDNA antibody single chain antibody fragment (scFv) libraries in phagemid vector pComb3X. Total RNA from uninfected individual PBMCs were pooled and mRNA was prepared from the pooled total RNA using the ‘Oligotex mRNA Mini Kit’ (Qiagen). The mRNA was used to reverse transcribe cDNA for construction of cDNA antibody libraries.
Construction of genomic DNA antibody scFv libraries
A total of 67 primers were designed and used in semi-nested PCR reactions to amplify heavy chain and kappa and lambda light chain variable regions using gDNA as a template, and in SOE-PCR to assemble scFvs for gDNA scFv library construction (Table S1). Reverse primers were mixed at equal molar concentrations prior to semi-nested PCRs. Each sense primer annealing to leader sequences or framework 1 (FR1) regions of heavy chain variable regions or kappa and lambda light chain variable regions was paired with the corresponding mixed antisense primers and used at the same final concentration (10 μmol/l) in the first or second round of PCRs. Each first round of PCR was carried out in a reaction volume of 100 μl containing 90 ng gDNA as a template by running the following PCR programme: initial denaturing at 95°C for 3 min, 10 cycles of 95°C for 15 s, 45°C for 30 s, 72°C for 45 s, 20 cycles of 95°C for 15 s, 55°C for 30 s, 72°C for 45 s and an extension cycle of 72°C for 10 min. The second round of PCR was carried out using the corresponding first round PCR product as a template and running the following PCR programme: initial denaturing at 95°C for 3 min, 30 cycles of 95°C for 15 s, 55°C for 30 s, 72°C for 30 s and an extension cycle of 72°C for 10 min. Semi-nested PCR products were gel-purified using a QIAquick gel extraction kit (Qiagen). The purified heavy chain variable regions and kappa and lambda light chain variable regions were then covalently linked by a flexible (G4S)3 linker by SOE-PCR to assemble scFvs using the primer pairs Sfi5new/LINKR and LINKF/Sfi3hisR. The SOE-PCR products were gel-extracted, digested with SfiI and ligated to pComb3X. The ligated products were then electroporated into TG1 electrocompetent cells.
Construction of cDNA antibody Fab libraries
The pooled mRNA from the healthy donors’ PBMCs and the mRNA from three elite controllers were used to reverse transcribe cDNA by using the ‘SuperScript III cDNA Synthesis Kit’. The cDNA was used as a template to amplify heavy chain variable regions and kappa/lambda light chains by PCR using high-fidelity DNA polymerase and primers annealing to FR1s and FR4s of human antibody heavy chain variable regions and primers annealing to FR1s and constant regions of human antibody light chains (Table S2). The human antibody heavy chain first constant domain (CH1) sequence was then attached to the heavy chain variable regions by SOE-PCR to assemble Fds. Light chains and Fds were further assembled by SOE-PCR to obtain Fab fragments. The Fab fragments were then ligated to pComb3X, and the ligated products electroporated into TG1 electrocompetent cells.
Deep sequencing and sequence analysis
Primers annealing to pComb3X vector or heavy and light chain constant regions (Table S1 and S2) were used to amplify the scFvs from the gDNA antibody libraries and Fds and light chains from the cDNA antibody libraries. Gel-purified PCR products were sent for deep sequencing using the Roche 454 genome sequencer FLX. Trim sequences (≥290 nt) were analysed using IMGT/HighV-QUEST software (http://imgt.org/HighV-QUEST/index.action).
Results
Construction of large combinatorial DNA and cDNA antibody libraries
We prepared the gDNAs from the PBMCs of 300 uninfected healthy donors and amplified the heavy chain variable regions and kappa and lambda light chain variable regions by semi-nested PCR using pooled gDNA as a template and a set of primers that annealed to the leader sequence or framework 1 (FR1) or FR4 of each heavy chain or kappa and lambda light chain gene family (Table S1). The heavy chain variable regions and kappa and lambda light chain variable regions were then covalently linked by a flexible (G4S)3 linker by strand overlap extension (SOE)-PCR to assemble scFvs. The resultant combinatorial gDNA scFv library, including two sublibraries, designated NIgH/gK and NIgH/gL, in pComb3X contained over 600 million individual clones. Similarly, we constructed three immune gDNA scFv libraries using the gDNAs isolated from pt1, pt2 and pt3. Each immune gDNA scFv library had kappa and lambda sublibraries except that pt1 lambda sublibrary was omitted. Five sublibraries were designated pt1gH/gK, pt2gH/gK, pt2gH/gL, pt3gH/gK and pt3gH/gL, and each sublibrary contained one to six billion individual clones. We simultaneously constructed corresponding combinatorial cDNA antibody Fab libraries except that construction of a cDNA Fab library using pt1 PBMCs was omitted because the isolation of bnmAbs by single cell sorting of memory B-cells from pt1 PBMCs has been reported [18,28]. The resultant nonimmune and immune cDNA Fab kappa and lambda sublibraries were designated NIH/NIK and NIH/NIL, pt2H/pt2K and pt2H/pt2L, and pt3H/pt3K and pt3H/pt3L, respectively. Each sublibrary contained 1–6 billion individual clones.
More diverse heavy chain V-gene lineage usage in the genomic DNA libraries than in the cDNA libraries
We amplified the scFvs from the gDNA libraries and the Fds and light chains from the cDNA libraries using primers that annealed to the pComb3X vector or to the constant regions of heavy or light chains (CH1 and CL, respectively) (Table S1 and S2) and sent the PCR products for deep sequencing. We obtained trim sequences (>290 nt) from each library ranging from 23 530 to 88 331 for heavy chain variable regions, and from 17 285 to 28 110 for kappa light chain variable regions and from 21 480 to 30 199 for lambda light chain variable regions. Sequence analysis showed different patterns of using various IGHV and IGKV/IGLV (kappa and lambda light chain V-genes) lineages in different gDNA and cDNA libraries, and the differences between the gDNA and corresponding cDNA libraries were more significant than those between the nonimmune and immune gDNA or cDNA libraries (Figs 1–3). The gDNA libraries were more diverse overall than the cDNA libraries in using various IGHV lineages (Figs 1 and 2). Among the four gDNA heavy chain libraries, NIgH and pt1gH showed a similar pattern of various IGHV lineage usage, whereas pt2gH and pt3gH were significantly different from NIgH and pt1gH in using IGHV1, IGHV2 and IGHV6 lineages (Figs 1 and 2). Compared with the gDNA heavy chain libraries, the corresponding cDNA heavy chain libraries had significantly higher percentages of clones using IGHV1 and IGHV3 lineages (Fig. 1), and were biased to certain VH1 and VH3 subfamilies, including IGHV1–18, 1–2 and 1–69, and IGHV 3–11, 3–21, 3–23, 3–30, 3–33, 3–49, 3–7 and 3–74 (Fig. 2). The patterns of various IGKV/IGLV lineage usages in the nonimmune and immune gDNA libraries were similar except for pt1gK library (Figs 1 and 3). The nonimmune and immune cDNA libraries also showed a similar pattern in using various IGKV/IGLV lineages. Both nonimmune and immune cDNA antibody libraries heavily used IGKV3 and IGLV1 lineages (Figs 1 and 3). These results indicate that HIV-1 infection shapes the patterns of various IGHV lineage usages, but the caused changes at the cDNA level are much less significant compared with the changes at the gDNA level. The differences between the gDNA and cDNA antibody gene repertoires in HIV-1 uninfected (nonimmune) humans reflect host immune regulations, and such regulations may largely determine the host-dependent immune response to HIV-1 infection.
Fig. 1.
Percentage of immunoglobulin heavy chain V-gene family and kappa/lambda light chain V-gene family in nonimmune and immune genomic DNA and cDNA antibody libraries.
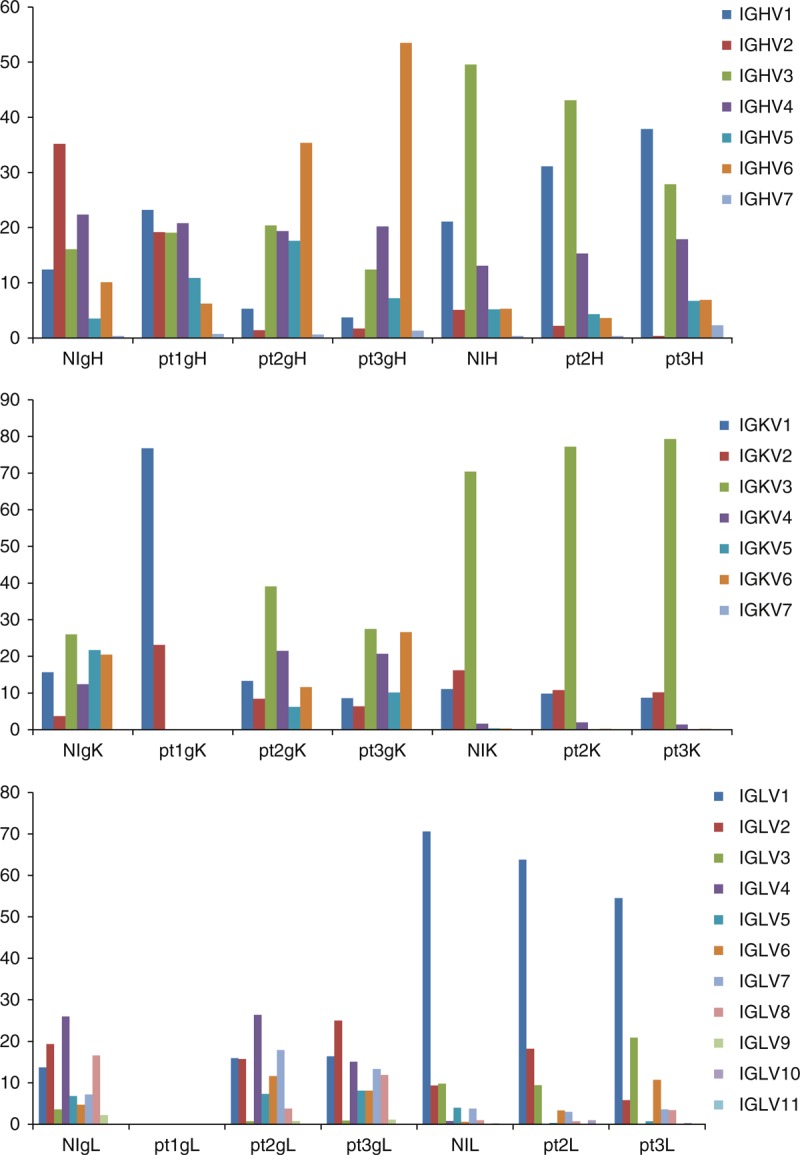
NIgH/K/L, nonimmune gDNA scFv library; NIH/K/L, nonimmune cDNA Fab library; pt1–3gH/K/L, patient gDNA scFv library; pt1–3H/K/L, patient cDNA Fab library. Note: pt1gL library is not available.
Fig. 2.
Percentage of heavy chain V-gene lineages in the nonimmune and immune genomic DNA and cDNA libraries.
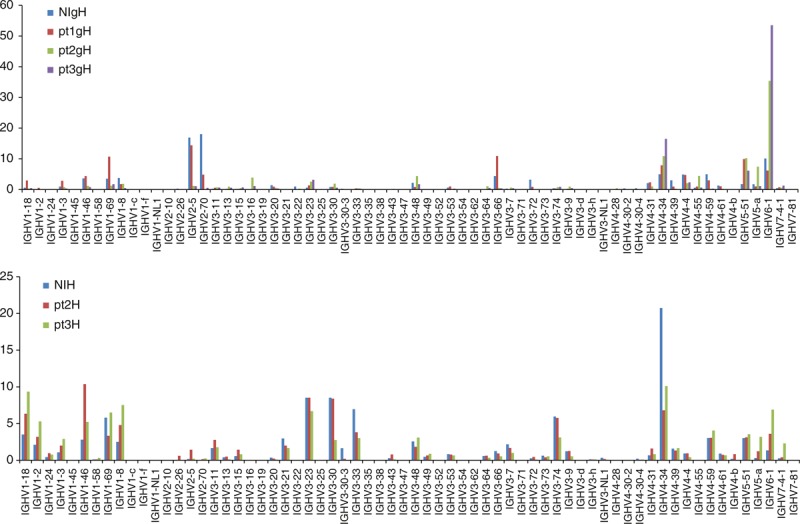
NIgH, nonimmune gDNA scFv library; NIH, nonimmune cDNA Fab library; pt1–3gH, patient gDNA scFv library; pt1–3H, patient cDNA Fab library.
Fig. 3.
Percentage of kappa light chain V-gene/lambda light chain V-gene lineages in the nonimmune and immune genomic DNA and cDNA libraries.
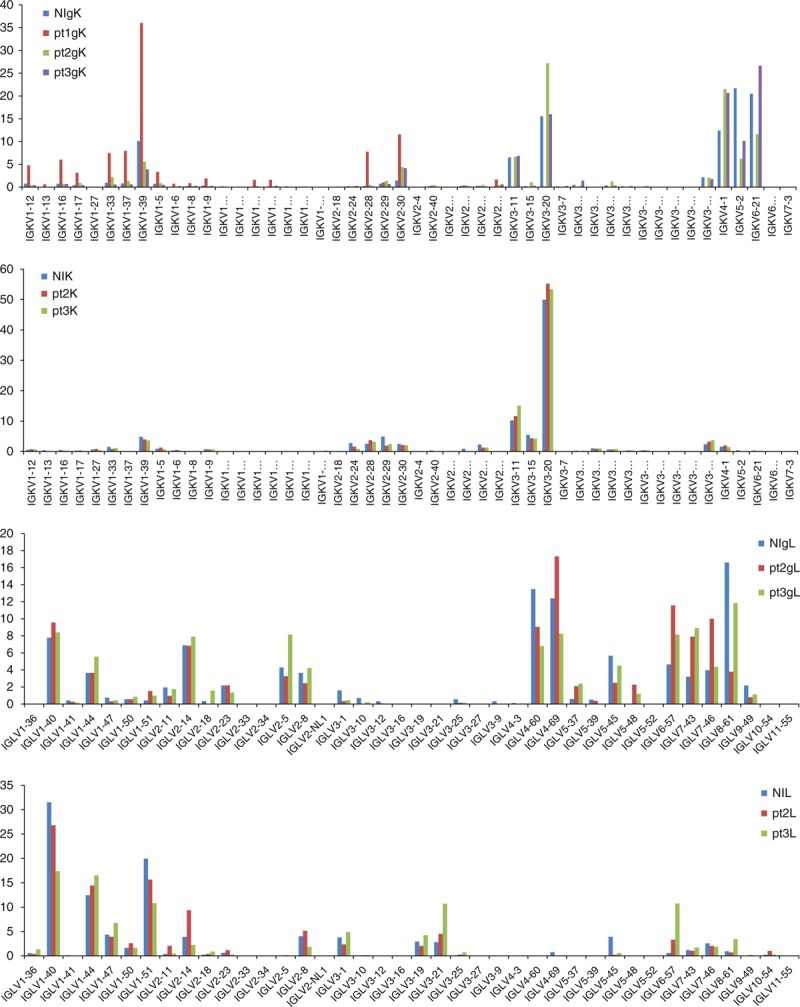
NIgK/L, nonimmune gDNA scFv library; NIK/L, nonimmune cDNA Fab library; pt1–3gK/L, patient gDNA scFv library; pt1–3K/L, patient cDNA Fab library.
Extremely low frequency of the heavy chain variable regions and kappa/lambda light chain variable regions with identical V(D)J recombinations to known HIV-1 bnmAbs
To investigate the potential of basal human antibody gene repertoires to develop HIV-1 bnAbs, we analysed the trim heavy chain variable region sequences and counted the number of heavy chain variable regions that had identical putative VDJ recombinations to five known CD4 binding site bnmAbs b12, VRC01, VRC03, NIH45–46 and 3BNC60, and two glycan and loop-specific bnmAbs PG9 and pGT127. To our surprise, we found that the frequencies of the heavy chain variable regions with identical putative VDJ recombinations to these known HIV-1 bnmAbs in both nonimmune and immune gDNA and cDNA libraries were extremely low (Table 1). We did not find any heavy chain variable regions that had identical VDJ recombinations compared with the known bnmAbs in the nonimmune and three immune gDNA libraries (Table 1). We found a total of five, 10 and two productive heavy chain variable regions (with in-frame junctions) in the cDNA libraries with exactly the same putative VDJ recombinations as VRC01, VRC03 and NIH45–46, respectively (Table 1). However, their junction regions and the length of the HCDR3s were very different compared with the respective bnmAbs (data not shown), suggesting the unlikelihood for them to mature to VRC01-like bnAbs. We did a similar search for heavy chain variable regions that had identical VDJ recombinations to a nonneutralizing or weakly neutralizing CD4-induced (CD4i) mAb X5 and a bnmAb against SARS-CoV, m396 [31,32]. We found four productive heavy chain variable regions in two immune gDNA libraries (1 in pt2gH and 3 in pt3gH) and a total of 28 productive heavy chain variable regions in the cDNA libraries (12 in NIH, 2 in pt2H and 14 in pt3H) that had identical putative VDJ recombinations to X5, but all these heavy chain variable regions had a shorter HCDR3 than that of X5 (24 amino acids, AA). We found one heavy chain variable region in pt1gH library and one heavy chain variable region in NIH library that had exactly the same putative VDJ recombination as m396, but none of them had the same HCDR3 length as that of m396 (11 amino acids). These results indicate that the chance of having a heavy chain variable region with a defined VDJ recombination along with a specific HCDR3 length could be extremely low in human antibody gene repertoires.
Table 1.
Number of productive heavy chain variable regions and kappa/lambda light chain variable regions with identical putative V(D)J recombinations to known HIV-1 bnmAbs.
| BnmAbs VHs | IGHV | IGHD | IGHJ | NIgH | pt1gH | pt2gH | pt3gH | NIH | pt2H | pt3H |
| b12 | HV1-3*01 | HD2-21*01 F | HJ6*03 F | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| VRC01 | HV1-2*02 | HD2-21*01 | HJ2*01 | 0 | 0 | 0 | 0 | 4 | 0 | 1 |
| VRC03 | HV1–2*02,04; HV1–8*01 | HD2–21*01 | HJ2*01 | 0 | 0 | 0 | 0 | 6 | 0 | 4 |
| NIH45–46 | HV1–2*02 | HD1-26 | HJ2*01 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| 3BNC60 | HV1–2*01 | HD3–3 | HJ2*01 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| PG9 | HV3–33*05 | HD1–1*01 F | HJ6*03 F | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| PGT127 | HV4–61*05 | HD3–16*02 | HJ5*02 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| X5 | HV1–69*01 | HD3–22*01 | HJ4*02 F | 0 | 0 | 1 | 3 | 12 | 2 | 14 |
| m396 | HV1–69*05 | HD5–18*01 | HJ6*02 F | 0 | 1 | 0 | 0 | 1 | 0 | 0 |
| Total no. of trim VH sequences | 33 436 | 23 530 | 44 027 | 42 160 | 80 331 | 74 053 | 77 176 | |||
| BnmAbs VKs/VLs | IGKV/IGLV | IGKJ/IGLJ | NIgK/L | pt1gK | pt2gK/L | pt3gK/L | NIK/L | pt2K/L | pt3K/L | |
| b12 | IGKV3-20 | IGKJ2 | 136 | 3 | 179 | 124 | 1172 | 1093 | 1280 | |
| VRC01 | IGKV3D-15 | IGKJ2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| VRC03 | IGKV3-NL5 | IGKJ2 | 0 | 0 | 0 | 0 | 1 | 5 | 0 | |
| NIH45–46 | IGKV3D-15 | IGKJ2 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | |
| 3BNC60 | IGKV1-33, IGKV1D-33 | IGKJ3 | 0 | 0 | 1 | 0 | 1 | 2 | 0 | |
| PG9 | IGLV2-14 | IGLJ3 | 181 | NA | 100 | 167 | 381 | 605 | 195 | |
| PGT127 | IGLV2–8 | IGLJ2, IGLJ3 | 90 | NA | 13 | 114 | 483 | 255 | 102 | |
| X5 | IGKV3-20 | IGKJ2 | 136 | 4 | 179 | 124 | 1172 | 1093 | 1280 | |
| m396 | IGLV3-21 | IGLJ1 | 0 | NA | 0 | 0 | 5 | 28 | 19 | |
| Total no. of trim VK and VL sequences | 23 670 | 21 254 | 19 162 | 17 285 | 26 902 | 26 508 | 27 639 | |||
CD4i mAb X5 and bnmAb m396 against SARS-CoV were included for comparison. VH, heavy chain variable region; VK, kappa light chain variable region; VL, lambda light chain variable region.
The frequencies of the kappa light chain variable regions that had the identical VJ recombinations to the known HIV-1 bnmAbs in both nonimmune and immune gDNA and cDNA libraries were also very low except that the frequencies of the kappa light chain variable regions with the identical VJ recombination to b12 kappa light chain variable region (IGKV3–20/IGKJ2) were relatively high (Table 1). X5 used the same VJ recombination as b12, so the frequencies of the kappa light chain variable regions with identical VJ recombination to X5 were also relatively high. Interestingly, we found that the frequencies of the lambda light chain variable regions with the identical VJ recombinations to bnmAbs PG9 and PGT127 were also relatively high, whereas the frequencies of the kappa light chain variable regions with the identical VJ recombination to m396 were very low (Table 1).
Relatively high frequencies of heavy and kappa/lambda light chain variable regions that used the same or very similar heavy chain genes and kappa/lambda light chain V-genes and had the same length of CDR3s as known HIV-1 bnmAbs regardless of (D)J-gene usage
It is usually difficult to determine which D-gene and J-gene were recombined with the V-genes to generate heavy chain variable regions, owing to the complexity of VDJ recombination events. Therefore, we searched for productive heavy chain variable regions that used the same or very similar IGHVs and had the same length of HCDR3 as known HIV-1 bnmAbs regardless of D-gene and J-gene usage. The frequencies of such heavy chain variables were relatively high in the nonimmune and immune gDNA and cDNA libraries than the frequencies of the heavy chain variable regions with exactly the same VDJ recombinations of known HIV-1 bnmAbs except for VRC03, PG9 and PGT127 that had long HCDR3s (23, 30 and 25 amino acids, respectively) (Table 2). X5 also had a long HCDR3 (24 amino acids), so the frequencies of the heavy chain variable regions that used the same or very similar IGHV of X5 and had the same HCDR3 length were not higher than the frequencies of the heavy chain variable regions with exactly the same VDJ recombinations of X5. Similar analysis of trim kappa/lambda light chain variable regions also showed relatively high frequencies of the VKs/VLs that used the same or very similar IGKVs/IGLVs and had the same length of CDR3 as known HIV-1 bnmAbs regardless of J-gene usage, especially for b12, PG9 and PGT127 (Table 2).
Table 2.
Number of heavy chain variable regions and kappa/lambda light chain variable regions that used the same or very similar V-genes and had the same CDR3 length as known HIV-1 bnmAbs.
| BnmAbs VHs | IGHVs | CDR3 length | NIgH | pt1gH | pt2gH | pt3gH | NIH | pt2H | pt3H |
| b12 | HV1–3*01,02;1–8*02;1–18*01,03 | 20 AA | 3 | 10 | 0 | 0 | 34 | 69 | 58 |
| VRC01 | HV1–2 *01-05 | 14 AA | 1 | 2 | 0 | 1 | 79 | 122 | 210 |
| VRC03 | HV1–2*02,04,05;1–8*01,02 | 23 AA | 0 | 2 | 0 | 0 | 8 | 4 | 58 |
| NIH45–46 | HV1–2*02–05;1–3*01 | 18 AA | 10 | 17 | 10 | 1 | 47 | 48 | 115 |
| 3BNC60 | HV1–2*01-05 | 17 AA | 0 | 7 | 0 | 0 | 37 | 24 | 227 |
| PG9 | HV3–33*01,05,06; &3–30*02,03 | 30 AA | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| PGT127 | HV4–39*03,06,07;4–61*05;4-b*02 | 25 AA | 0 | 1 | 0 | 0 | 1 | 2 | 0 |
| X5 | HV1–69*01,05,06,12,13 | 24 AA | 2 | 3 | 0 | 2 | 3 | 3 | 4 |
| m396 | HV1–69*01,02,04,05,09 | 11 AA | 5 | 18 | 2 | 7 | 77 | 42 | 38 |
| Total no. of trim VH sequences | 33 436 | 23 530 | 44 027 | 42 160 | 80 331 | 74 053 | 77 176 | ||
| BnmAbs VKs/VLs | IGKVs/IGLVs | CDR3 length | NIgK/L | pt1gK | pt2gK/L | pt3gK/L | NIK/L | pt2K/L | pt3K/L |
| b12 | IGKV3–20, 3D-20, 3-NL5, 3D-15 | 9 AA | 177 | 5 | 218 | 159 | 1834 | 1665 | 2117 |
| VRC01 | IGKV3–15, 3D-15, 3–7 | 7 AA | 1 | 0 | 0 | 1 | 0 | 0 | 1 |
| VRC03 | IGKV3-NL5, 3–20, 3D-20, 3D-15 | 5 AA | 1 | 0 | 5 | 0 | 7 | 10 | 6 |
| NIH45–46 | IGKV3–15, 3D-15, 3–7 | 5 AA | 0 | 0 | 1 | 1 | 1 | 4 | 0 |
| 3BNC60 | IGKV1–33, 1D-33, 1–39, 1D-39 | 5 AA | 0 | 14 | 1 | 0 | 1 | 5 | 0 |
| PG9 | IGLV2–14, 2-23 | 10 AA | 377 | NA | 266 | 320 | 675 | 1201 | 298 |
| PGT127 | IGLV2–8, IGLV2-11 | 10 AA | 143 | 26 | 179 | 522 | 343 | 142 | |
| X5 | KV3–20, KV3D-20, KV3-NL5, KV3D-7 | 9 AA | 181 | 5 | 218 | 155 | 1805 | 1655 | 2103 |
| m396 | IGLV3–21, IGLV3–9 | 10 AA | 9 | NA | 0 | 0 | 61 | 141 | 230 |
| Total no. of trim VK and VL sequences | 23 670 | 21 254 | 19 162 | 17 285 | 26 902 | 26 508 | 27 639 | ||
CD4i mAb X5 and bnmAb m396 against SARS-CoV were included for comparison. VH, heavy chain variable; VK, kappa light chain variable; VL, lambda light chain variable.
Discussion
B-cell antigen receptors (BCRs) develop as B-cells differentiate. VDJ recombination for the heavy chain and VJ recombination for the light chain occur sequentially at pro-B and pre-B stages, respectively, in the bone marrow. Immature B-cells exit the bone marrow and enter the peripheral system in which new emigrant B cells differentiate into immature and then mature naive B-cells. Mature B-cells undergo somatic maturation upon immunogen stimulation and differentiate into Ab-secreting plasma cells or memory B-cells. Thus, the peripheral blood contains a population of B-cells that have undergone V(D)J recombinations, and the gDNAs of peripheral B-cells of nonimmune humans possess the basal rearranged antibody gene repertoire for antibody affinity maturation. The genome-based antibody gene repertoire may differ from the cDNA-based antibody gene repertoire owing to the different transcriptional and/or translational levels of different antibody genes, and both repertoires may be shaped upon viral infection or vaccination. Therefore, we constructed a large combinatorial nonimmune and three immune human gDNA antibody libraries and their corresponding cDNA antibody libraries for comparison of antibody gene repertoires at the gDNA and cDNA levels by deep sequencing and for subsequent isolation of Env-specific germline or intermediate antibodies (with a low level of somatic maturation) by phage display. We found that frequencies of the heavy chain variable regions and kappa/lambda light chain variable regions with identical putative V(D)J recombinations to known HIV-1 bnmAbs were extremely low in both nonimmune gDNA and cDNA libraries, suggesting that known HIV-1 bnmAbs may not be derived from the putative germline antibodies, or the chance for such direct maturation may be very low. If a certain combination of putative germline heavy and light chains is required for HIV-1 bnAbs, the frequency of such germline antibody genes may be even lower. Compared with the lack of measurable binding of putative germline antibodies of known HIV-1 bnmAbs to Env and the requirement of extensive somatic maturation for broadly neutralizing activity [13,27], limited availability of B-cells bearing proper germline antibody genes that may mature to HIV-1 bnAbs upon stimulation may be a formidable challenge for developing an effective HIV-1 vaccine. The extremely low frequency of the putative germline antibody genes of known HIV-1 bnmAbs in the human antibody gene repertoires suggests that the approach for eliciting HIV-1 bnAbs by identifying primary immunogens to trigger the putative germline antibodies of known HIV-1 bnmAbs in vivo may have a very limited chance of success. However, our further sequence analyses revealed relatively high frequencies of the heavy chain variable regions and kappa/lambda light chain variable regions in the human antibody gene repertoires that use the same or very similar IGHVs and IGKVs/IGLVs and have the same CDR3 length as known HIV-1 bnmAbs regardless of (D)J-gene usage. B-cells harbouring such heavy chain variable regions and kappa/lambda light chain variable regions may be stimulated to induce bnAbs. Note that we used the plasmids from the cDNA Fab libraries and the gDNA scFv libraries as templates for PCR amplification of the heavy chain variable regions and kappa/lambda light chain variable regions for deep sequencing. The cDNA Fab libraries were constructed earlier and used in our previous studies. We did not convert the Fab libraries to scFv libraries because it was not necessary to do so. Conversion between two different formats of antibody libraries may cause loss of antibody diversity and result in bias to certain antibody gene families.
Exploring the human gDNA antibody gene repertoire may be another approach for eliciting HIV-1 bnAbs. As we hypothesized, the gDNA antibody gene repertoires showed more diverse usages of IGHV lineages than the cDNA repertoires. Indeed, we have isolated a panel of RSC3-specific antibodies from our combinatorial gDNA libraries, but not from the cDNA libraries (manuscript in preparation). The isolated RSC3-specific antibodies had no or very low levels of somatic maturation and used diverse V(D)J recombinations, but they competed with mature b12 and VRC01 for binding to engineered Env, RSC3 [13], suggesting their potential to mature to VRC01-like bnAbs. We are currently doing in-vitro maturation and selection to confirm that they can potentially mature to VRC01-like bnAbs.
We obtained 33 436 and 80 331 trim heavy chain variable region sequences from 454 deep sequences of the nonimmune gDNA and cDNA libraries, respectively (Tables 1 and 2), which were comparable to the theoretical maximum diversity of the basal human heavy chain variable gene repertoire (3.1 × 104). The unequal usage of IGHV, IGHD and IGHJ genes in the nonimmune gDNA library suggests that VDJ recombination may not be a random event. The differences may be amplified by host immune regulations to remove or functionally silence B-cells expressing autoimmune antibodies or to deplete the B-cells that recognize B-cell superantigens [33–35]. This could lead to a different frequency of various germline antibody genes in the initial antibody gene repertoire for affinity maturation. For example, the heavy chain variable regions using IGHV1–2 or with HCDR3s of long length (20 amino acids and over) were significantly less frequent than the heavy chain variable regions using IGHV1–46 or with HCDR3s of medium length (10–15 amino acids) (Fig. 2 and data not shown). Arnaout et al.[36] also showed that V, D and J segments were utilized with different frequencies, resulting in a highly skewed representation of VDJ combinations in the human antibody gene repertoire. However, they found that the pattern of segment usage was almost identical between two different individuals. Our result seems different from their finding. We found that IGHV lineage usage differs from individual to individual. In addition, they reported that IGHV1–2 lineage accounted for 2–3% sequence in nonimmune human antibody gene repertoire [36]. We found a similar percentage in our cDNA libraries, but the percentages of IGHV1–2 lineage in gDNA libraries were 10 to 100-fold lower (Fig. 2). The more diverse usages of IGHV lineages in the gDNA libraries may account for this phenomenon.
Our study suggests the potential of the human immune system to develop HIV-1 bnAbs, which may have implications for vaccine development. In addition to searching for proper germline antibodies as targets for vaccine immunogen design, immune modulations may be required to tackle possible obstacles posed by the host and/or the virus to affinity maturation to HIV-1 bnAbs.
Acknowledgements
We wish to thank David D. Ho for the patient 1 sample, Zhi-Yong Yang, Tongqing Zhou and Peter D. Kwong for RSC3 and ΔRSC3 proteins, Xueling Wu and John R. Mascola for VRC01 antibody, Dennis R. Burton for the pComb3X plasmid, Peter D. Kwong, John R. Mascola, Tongqing Zhou, Xueling Wu and Dimiter S. Dimitrov for helpful discussions, and Jenny Ng for editing the manuscript.
M-Y. Z. designed research; Y.Z., T.Y., J.L. Y.Z. and M-Y. Z. performed research; Y.Z., Y.Z. and M-Y.Z. analysed data; Z.C. and Y.S. contributed patient samples; and M-Y.Z. and J.X. wrote the article.
This work was supported by Hong Kong Research Grants Consuls (RGC) General Research Fund (GRF) (#785210 and #785112) and China 12th 5-year Mega project for HIV/AIDS (#2012ZX10001006) to M-Y.Z. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Conflicts of interest
There are no conflicts of interest.
Supplementary Material
Footnotes
Correspondence to Mei-Yun Zhang, PhD, AIDS Institute, Department of Microbiology, Li Ka Shing Faculty of Medicine, The University of Hong Kong. L5-45, Laboratory Block, 21 Sassoon Road, Pokfulam, Hong Kong, China. Tel: +852 28183685; fax: +852 28177805; e-mail: zhangmy@hku.hk
References
- 1.Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 1998; 393:648–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wyatt R, Kwong PD, Desjardins E, Sweet RW, Robinson J, Hendrickson WA, et al. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature 1998; 393:705–711 [DOI] [PubMed] [Google Scholar]
- 3.Kwong PD, Wyatt R, Sattentau QJ, Sodroski J, Hendrickson WA. Oligomeric modeling and electrostatic analysis of the gp120 envelope glycoprotein of human immunodeficiency virus. J Virol 2000; 74:1961–1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kwong PD, Doyle ML, Casper DJ, Cicala C, Leavitt SA, Majeed S, et al. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature 2002; 420:678–682 [DOI] [PubMed] [Google Scholar]
- 5.Shaw GM, Hahn BH, Arya SK, Groopman JE, Gallo RC, Wong-Staal F. Molecular characterization of human T-cell leukemia (lymphotropic) virus type III in the acquired immune deficiency syndrome. Science 1984; 226:1165–1171 [DOI] [PubMed] [Google Scholar]
- 6.Hahn BH, Gonda MA, Shaw GM, Popovic M, Hoxie JA, Gallo RC, et al. Genomic diversity of the acquired immune deficiency syndrome virus HTLV-III: different viruses exhibit greatest divergence in their envelope genes. Proc Natl Acad Sci U S A 1985; 82:4813–4817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wong-Staal F, Shaw GM, Hahn BH, Salahuddin SZ, Popovic M, Markham P, et al. Genomic diversity of human T-lymphotropic virus type III (HTLV-III). Science 1985; 229:759–762 [DOI] [PubMed] [Google Scholar]
- 8.Burton DR, Pyati J, Koduri R, Sharp SJ, Thornton GB, Parren PW, et al. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 1994; 266:1024–1027 [DOI] [PubMed] [Google Scholar]
- 9.Purtscher M, Trkola A, Gruber G, Buchacher A, Predl R, Steindl F, et al. A broadly neutralizing human monoclonal antibody against gp41 of human immunodeficiency virus type 1. AIDS Res Hum Retroviruses 1994; 10:1651–1658 [DOI] [PubMed] [Google Scholar]
- 10.Scanlan CN, Pantophlet R, Wormald MR, Ollmann Saphire E, Stanfield R, Wilson IA, et al. The broadly neutralizing antihuman immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of alpha1-->2 mannose residues on the outer face of gp120. J Virol 2002; 76:7306–7321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stiegler G, Kunert R, Purtscher M, Wolbank S, Voglauer R, Steindl F, et al. A potent cross-clade neutralizing human monoclonal antibody against a novel epitope on gp41 of human immunodeficiency virus type 1. AIDS Res Hum Retroviruses 2001; 17:1757–1765 [DOI] [PubMed] [Google Scholar]
- 12.Scheid JF, Mouquet H, Feldhahn N, Walker BD, Pereyra F, Cutrell E, et al. A method for identification of HIV gp140 binding memory B cells in human blood. J Immunol Methods 2009; 343:65–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 2010; 329:856–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corti D, Langedijk JP, Hinz A, Seaman MS, Vanzetta F, Fernandez-Rodriguez BM, et al. Analysis of memory B cell responses and isolation of novel monoclonal antibodies with neutralizing breadth from HIV-1-infected individuals. PLoS ONE 2010; 5:e8805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP, et al. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 2011; 477:466–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, et al. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 2009; 326:285–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu X, Zhou T, Zhu J, Zhang B, Georgiev I, Wang C, et al. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science 2011; 333:1593–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scheid JF, Mouquet H, Ueberheide B, Diskin R, Klein F, Oliveira TY, et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science 2011; 333:1633–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang J, Ofek G, Laub L, Louder MK, Doria-Rose NA, Longo NS, et al. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature 2012; 491:406–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Calarese DA, Lee HK, Huang CY, Best MD, Astronomo RD, Stanfield RL, et al. Dissection of the carbohydrate specificity of the broadly neutralizing anti-HIV-1 antibody 2G12. Proc Natl Acad Sci U S A 2005; 102:13372–13377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McLellan JS, Pancera M, Carrico C, Gorman J, Julien JP, Khayat R, et al. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature 2011; 480:336–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pancera M, McLellan JS, Wu X, Zhu J, Changela A, Schmidt SD, et al. Crystal structure of PG16 and chimeric dissection with somatically related PG9: structure-function analysis of two quaternary-specific antibodies that effectively neutralize HIV-1. J Virol 2010; 84:8098–8110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pejchal R, Doores KJ, Walker LM, Khayat R, Huang PS, Wang SK, et al. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science 2011; 334:1097–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saphire EO, Parren PW, Pantophlet R, Zwick MB, Morris GM, Rudd PM, et al. Crystal structure of a neutralizing human IGG against HIV-1: a template for vaccine design. Science 2001; 293:1155–1159 [DOI] [PubMed] [Google Scholar]
- 25.Zhou T, Georgiev I, Wu X, Yang ZY, Dai K, Finzi A, et al. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science 2010; 329:811–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou T, Xu L, Dey B, Hessell AJ, Van Ryk D, Xiang SH, et al. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature 2007; 445:732–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiao X, Chen W, Feng Y, Zhu Z, Prabakaran P, Wang Y, et al. Germline-like predecessors of broadly neutralizing antibodies lack measurable binding to HIV-1 envelope glycoproteins: implications for evasion of immune responses and design of vaccine immunogens. Biochem Biophys Res Commun 2009; 390:404–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scheid JF, Mouquet H, Feldhahn N, Seaman MS, Velinzon K, Pietzsch J, et al. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature 2009; 458:636–640 [DOI] [PubMed] [Google Scholar]
- 29.Bonsignori M, Montefiori DC, Wu X, Chen X, Hwang KK, Tsao CY, et al. Two distinct broadly neutralizing antibody specificities of different clonal lineages in a single HIV-1-infected donor: implications for vaccine design. J Virol 2012; 86:4688–4692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu X, Hong K, Zhao C, Zheng Y, Ma L, Ruan Y, et al. Profiles of neutralizing antibody response in chronically human immunodeficiency virus type 1 clade B’-infected former plasma donors from China naive to antiretroviral therapy. J Gen Virol 2012; 93:2267–2278 [DOI] [PubMed] [Google Scholar]
- 31.Zhu Z, Chakraborti S, He Y, Roberts A, Sheahan T, Xiao X, et al. Potent cross-reactive neutralization of SARS coronavirus isolates by human monoclonal antibodies. Proc Natl Acad Sci U S A 2007; 104:12123–12128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moulard M, Phogat SK, Shu Y, Labrijn AF, Xiao X, Binley JM, et al. Broadly cross-reactive HIV-1-neutralizing human monoclonal Fab selected for binding to gp120-CD4-CCR5 complexes. Proc Natl Acad Sci U S A 2002; 99:6913–6918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zouali M. B cell superantigens subvert innate functions of B cells. Chem Immunol Allergy 2007; 93:92–105 [DOI] [PubMed] [Google Scholar]
- 34.Paul S, Planque SA, Nishiyama Y, Hanson CV, Massey RJ. Nature and nurture of catalytic antibodies. Adv Exp Med Biol 2012; 750:56–75 [DOI] [PubMed] [Google Scholar]
- 35.Planque SA, Mitsuda Y, Nishiyama Y, Karle S, Boivin S, Salas M, et al. Antibodies to a superantigenic glycoprotein 120 epitope as the basis for developing an HIV vaccine. J Immunol 2012; 189:5367–5381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arnaout R, Lee W, Cahill P, Honan T, Sparrow T, Weiand M, et al. High-resolution description of antibody heavy-chain repertoires in humans. PLoS ONE 2011; 6:e22365. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


