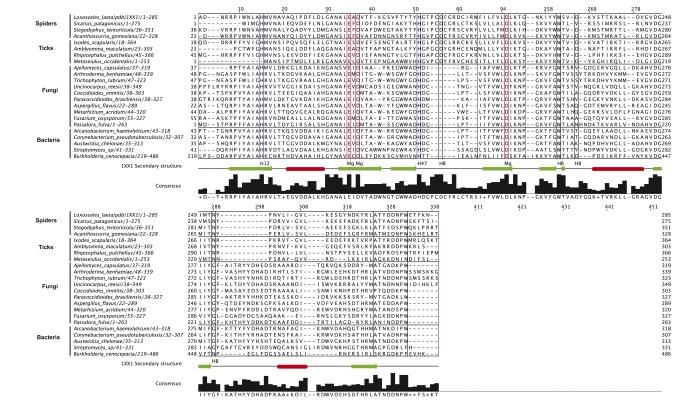
Figure 2. Multiple sequence alignment of one SMase D sequence for each genus identified.
Consensus sequences and secondary structure of L. laeta SMase D (pdb number:1xx1) are shown underneath the alignment. Conserved catalytic histidines residues are marked with blue boxes (Label H12 and H47 in secondary structure of 1xx1). Residues responsible for Mg+2 coordination are marked with red boxes (Label Mg in secondary structure of 1xx1) and amino acids involved in network of hydrogen bonds are marked with black boxes (label HB in secondary structure of 1xx1). The following sequences were used to build this alignment and were obtained from the NCBI nr protein database: Ajellomyces (GI: 225556466), Arthroderma (GI: 302499957), Trichophyton (GI: 327309460), Uncinocarpus (GI: 258566630), Coccidioides (GI: 119182286), Paracoccidioides (GI: 225681750), Aspergillus (GI: 220691453), Metaseiulus (GI: 391334832), Loxosceles (GI: 60594084), Sicarius (GI: 292630576), Ixodes (GI: 121962650), Amblyomma (GI: 346467405), Rhipicephalus (GI: 427782159), Burkholderia (GI: 116687281), Arcanobacterium (GI: 297571658), Corynebacterium (GI: 300857446), Streptomyces (GI: 254384004) and Austwickia (GI:403190737). Six genera have not entries in NCBI nr protein database, but their protein sequences were translated from in NCBI WGS or dbEST entries:, Fusarium (GI: 144921672, position: 31242-32224), Gibberella (GI: 116139506, position: 93710-94513), Passalora (GI: 407486978, position: 4015-4803) , Metarhizium (GI: 322696462), Stegodyphus (GI: 374247203) and Acanthoscurria (GIs: 68762186 and 68761727). The genera Dermatophagoides, Psoroptes, Varroa and Tetranycus have incomplete sequences and were not used in this analysis.