Figure 4.
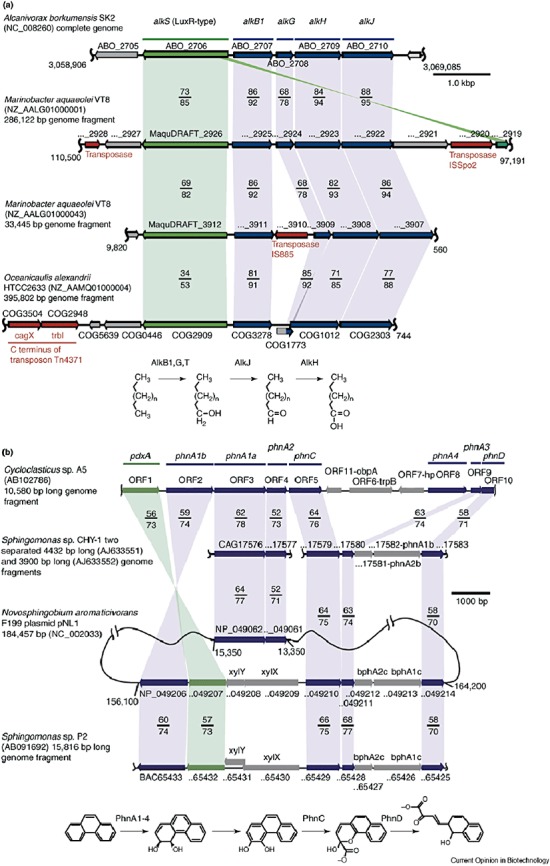
Ubiquity of gene clusters for the degradation of (A) aliphatic and (B) (poly) aromatic fractions of oil. A. Organization of genes homologous to the A. borkumensis alk gene cluster in the hydrocarbon‐degrading marine proteobacteria. Homologous genes are highlighted by shaded areas: sequences predicted to code for LuxR‐type transcriptional activators of the alkane genes, AlkS, are marked in green, genes for the alkane degradation pathway are indicated in blue. Percentages of protein identity/similarity of polypeptides from A. borkumensis with those of M. aqaeolei and O. alexandrii are shown. Gene designations: alkB1, alkane monooxygenase; alkG, rubredoxin; alkJ, alcohol dehydrogenase; alkH, aldehyde dehydrogenase. B. Organization of gene clusters of polycyclic aromatic hydrocarbons (PAH) degradation pathways of Cycloclasticus sp. A5 and PAH‐degrading α‐proteobacteria. pdxA genes predicted to code for the pyridoxal‐phosphate biosynthesis enzyme are colored in green, PAH degradation genes are in blue. Percentages of protein identity/similarity of the polypeptides from Cycloclasticus sp. A5 with those of the PAH‐degrading α‐proteobacteria are shown. Gene designations: phnA1, iron‐sulfur protein (ISP) α subunit of PAH dioxygenase; phnA2, ISP β‐subunit of PAH dioxygenase; phnA3, Rieske‐type [2Fe‐2S] ferredoxin, phnA4, NADH‐ferredoxin oxidoreductase; phnC, extradiol [3,4‐dihydrophenanthrene] dioxygenase; phnD, 2‐hydroxy‐2H‐benzo[h]chromene‐2‐carboxylate isomerase. Reprinted from Yakimov and colleagues (2007) with permission from Elsevier.
