Summary
Plant‐parasitic nematodes are the most destructive group of plant pathogens worldwide and are extremely challenging to control. Some Bacillus thuringiensis crystal proteins are highly toxic to the plant‐parasitic nematode Meloidogyne incognita. In this study, the nematicidal crystal proteins Cry6Aa, Cry5Ba and Cry55Aa were tested against M. incognita to select the best toxin combination for its management. The results showed that a combination of Cry6Aa and Cry55Aa showed significant synergistic toxicity against M. incognita, and the highest synergistic effect (five times the expected toxicity of the two toxins calculated from their separate toxicities) was observed when they were combined in a 1:1 ratio. Furthermore, ligand blot analyses of the interaction between total proteins of M. incognita and the three toxins showed many different signal bands, indicating that there is a range of host proteins with which the toxins can interact. One explanation of the observed synergism is that the toxins damage the host in diverse ways, and they may thus act cooperatively and thereby show greater toxicity in combination. Our discovery provides an effective strategy for controlling M. incognita by using a combination of Cry6Aa and Cry55Aa.
Introduction
Root‐knot nematodes can cause much damage to a broad range of crops, resulting in drastic yield losses, mainly in tropical and subtropical agriculture (Trudgill and Blok, 2001; Bird et al., 2009). Root‐knot nematodes are difficult to control because of their endoparasitic life style. They live underground and spend most of their lives in the roots, and the roots can protect them even from chemical nematicides. Chemical nematicides are still by far the most common current means of controlling root‐knot nematodes, but their availability is decreasing because of their toxicity to humans and the environment (Trudgill and Blok, 2001; Bird et al., 2009).
Bacillus thuringiensis is a rod‐shaped, Gram‐positive, spore‐forming bacterium with characteristic parasporal crystals formed during the stationary phase of growth (Schnepf et al., 1998; Roh et al., 2007). These crystals comprise one or more insecticidal crystal proteins that are toxic to many insect orders, such as Lepidoptera, Diptera and Coleoptera (Bravo et al., 2007; van Frankenhuyzen, 2009; Ohba et al., 2009). Given its strong and specific toxicity to a wide range of insects, B. thuringiensis has been developed as the leading biopesticide for use as an alternative or supplement to synthetic chemical pesticides (Rosas‐Garcia, 2009). In addition, its cry genes have also been considered a key to generating transgenic crops with pest resistance (Kumar et al., 2008; Shu and Zhang, 2009).
In the past few decades, several studies of the nematicidal effects of B. thuringiensis crystal proteins have been reported. Griffitts and colleagues (2001) reported that Cry5B, Cry6A and Cry14A led to lower brood size in Caenorhabditis elegans. Wei and colleagues (2003) reported that Cry6A, Cry5B, Cry21A and Cry14A are toxic to multiple nematode species, such as C. elegans, Panagrellus redivivus and Distolabrellus veechi. Li and colleagues (2007) reported that Cry6A expression in transgenic roots significantly impaired the ability of Meloidogyne incognita to reproduce. In addition, expression of a truncated 79 kDa version of Cry5B in transgenic roots reduced the number of galls produced by M. incognita and reduced the number of progeny by nearly threefold (Li et al., 2008). Previously, our group also isolated some B. thuringiensis strains that showed high levels of activity against plant‐parasitic nematodes (Guo et al., 2008; Yu et al., 2008). In these studies, three nematicidal crystal protein genes, cry6Aa, cry5Ba and cry55Aa, were cloned from the highly nematicidal B. thuringiensis strain YBT‐1518. Bioassays showed that these three crystal proteins were highly toxic to second‐stage juveniles (J2) of Meloidogyne hapla when applied in soluble form (Guo et al., 2008).
Plant‐parasitic nematodes usually live underground, which makes them difficult to target using traditional B. thuringiensis insecticides. One of the most effective approaches for controlling plant‐parasitic nematodes has been constructing transgenic plants with nematicidal cry genes. Either cry6A or cry5B alone expressed in the tomato provided good protection against M. incognita (Li et al., 2007). Transgenic crops producing two or more different B. thuringiensis toxins targeting the same plant‐parasitic nematode would probably be able to control nematodes more effectively. In this study, three nematicidal crystal proteins, Cry6Aa, Cry5Ba and Cry55Aa, were combined in an assay of their toxicities against M. incognita, with the aim of selecting the best toxin combination for M. incognita management and to understand the possible mechanism of synergism.
Results and discussion
Bioassay results of M. incognita J2 with Cry5Ba, Cry6Aa and Cry55Aa toxins, alone and in combinations (1:1), are summarized in Table 1. These show that Cry55Aa [50% lethal concentration (LC50) 102.57 µg ml−1 (95% fiducial limits determined by probit analysis 76.33–137.84) µg ml−1] has similar toxicity to Cry5Ba [LC50 146.05 (114.98–185.51) µg ml−1]. Cry5Ba was about twice as toxic as Cry6Aa [LC50 383.42 (266.54–551.57) µg ml−1]. When bioassays were performed with toxin mixture (1:1), the Cry6Aa‐Cry55Aa combination [LC50 32.14 (24.60–41.97) µg ml−1] showed a significant synergistic effect, about five times the reduction in LC50 value when compared with expected value (LC50 160.65 µg ml−1; we refer to this as a synergistic factor of 5). The toxin mix (1:1) of Cry6Aa and Cry5Ba was also about twice as toxic to M. incognita J2 than expected. However, the Cry5Ba‐Cry55Aa and Cry6Aa‐Cry5Ba‐Cry55Aa toxin mixtures showed no significant synergistic effect in toxicity towards M. incognita J2.
Table 1.
Effect of Bacillus thuringiensis nematicidal toxin and toxin mixtures (1:1) on mortality of Meloidogyne incognita J2.
| Toxins | Toxicity (µg ml−1) | Synergistic factor | |
|---|---|---|---|
| LC50a (observed) | LC50 (expected)b | ||
| Cry6Aa | 383.42 (266.54–551.57) | – | – |
| Cry55Aa | 102.57 (76.33–137.84) | – | – |
| Cry5Ba | 146.05 (114.98–185.51) | – | – |
| Cry6Aa‐Cry55Aa | 32.14 (24.60–41.97) | 160.65 | 5.00 |
| Cry6Aa‐Cry5Ba | 108.54 (80.29–146.74) | 211.52 | 1.95 |
| Cry55Aa‐Cry5Ba | 111.54 (83.72–148.60) | 121.37 | 1.09 |
| Cry6Aa‐Cry55Aa‐Cry5Ba | 127.39 (87.65–185.14) | 158.00 | 1.24 |
95% fiducial limits determined by probit analysis are given in parentheses.
Expected LC50 values were calculated using the equation of Tabashnik (1992).
Because the Cry6Aa‐Cry55Aa combination showed the greatest synergistic effect, experiments were carried out with different ratios of these toxins (1:5, 1:2, 1:1, 2:1, and 5:1; Table 2). The results showed that with all combinations of Cry1Ab and Cry1Ac tested, a decrease in LC50 values (ranging from 32.14 to 74.29 µg ml−1) when compared with the expected values were observed. When the ratio of Cry6Aa to Cry55Aa was 1:1, the toxicity to M. incognita was greatest (32.14 µg ml−1) and the calculated synergistic factor was 5.0. Even at a 5:1 ratio, a synergistic factor of about 3.3 was observed.
Table 2.
Bioassay of different ratios of Cry6Aa‐Cry55Aa toxin mixtures against Meloidogyne incognita J2.
| Cry6Aa : Cry55Aa | Toxicity (µg ml−1) | Synergistic factor | |
|---|---|---|---|
| LC50a (observed) | LC50 (expected)b | ||
| 1:0 | 383.42 (266.54–551.57) | – | – |
| 0:1 | 102.57 (76.33–137.84) | – | – |
| 1:5 | 49.43 (38.10–64.14) | 119.91 | 2.43 |
| 1:2 | 37.45 (22.37–74.66) | 135.27 | 3.61 |
| 1:1 | 32.14 (24.60–41.97) | 160.65 | 5.00 |
| 2:1 | 50.87 (35.17–72.55) | 201.42 | 3.96 |
| 5:1 | 74.29 (58.89–93.73) | 243.31 | 3.28 |
95% fiducial limits determined by probit analysis are given in parentheses.
Expected LC50 values were calculated using the equation of Tabashnik (1992).
Protein structure analysis was used to predict the mode of action of Cry5Ba, Cry6Aa and Cry55Aa. A phylogeny of the Cry toxins that have been documented to be toxic to nematodes is shown in Fig. S1. Cry6Aa, Cry5Ba and Cry55Aa belong to three different subfamilies (Fig. S1). Of the five blocks in the N‐terminal region of crystal proteins that are conserved between most of the family, Cry1A and Cry5Ba share three blocks and Cry6Aa and Cry55Aa share none (Fig. S2). In addition, the interaction between toxins and M. incognita were analysed by ligand blotting to understand the range of host proteins with which Cry5Ba, Cry6Aa and Cry55Aa can interact. This revealed positive signal bands for all three crystal proteins, indicating that all three could bind to host proteins. However, the binding protein signal profiles were very different between Cry5Ba, Cry6Aa and Cry55Aa (Fig. 1). The different protein structures and binding patterns of the three proteins with M. incognita proteins indicates that the modes of action of Cry6Aa, Cry5Ba and Cry55Aa in targeting M. incognita may be different.
Figure 1.
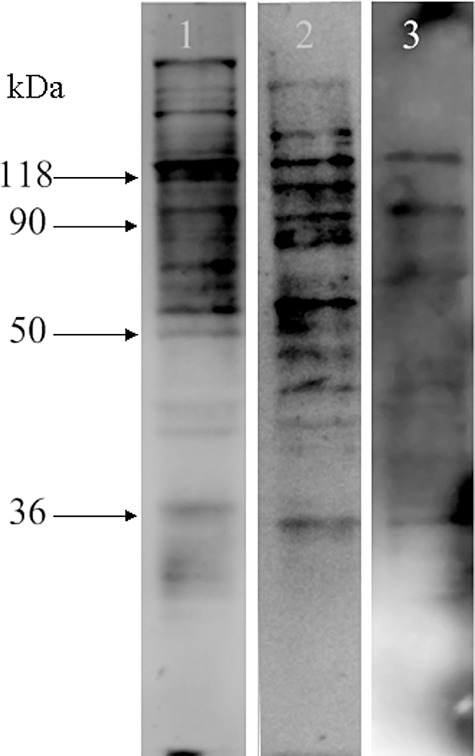
Analysis of the interaction between total proteins of M. incognita and nematicidal toxins Cry5Ba (lane 1), Cry6Aa (lane 2) and Cry55Aa (lane 3). The ability of Cry toxins to bind to M. incognita is shown on a nitrocellulose membrane and detected by antibodies against the respective toxin.
The use of synergism and combination of toxins has become one of the most effective methods for increasing the toxicity of B. thuringiensis biopesticides. Various hypotheses have been proposed to explain the molecular mechanism of synergism, including (i) improvement of toxin docking and membrane insertion (Perez et al., 2005) and (ii) destruction of the midgut peritrophic matrix to increase toxin permeability (Sampson and Gooday, 1998; Fang et al., 2009). In this study, we found that the nematicidal crystal protein combination Cry6Aa‐Cry55Aa showed clear synergistic toxicity against M. incognita, and the highest toxicity could be observed when the two proteins were present at a 1:1 ratio. We also found that there may be different modes of action among Cry6Aa, Cry5Ba and Cry55Aa when targeting M. incognita, because of their different protein structures and their different binding patterns to M. incognita proteins. An explanation of the observed synergism is that the toxins damage the host in the different ways and may thus act cooperatively and show higher toxicity to M. incognita in combination. Understanding the detailed mechanism of the observed synergism requires further experimentation.
This study demonstrated that the toxin combinations Cry6Aa‐Cry55Aa and Cry6Aa‐Cry5Ba showed clear synergistic toxicity against M. incognita. This knowledge could be useful when designing B. thuringiensis toxin expression strategies in transgenic plants. Plant promoters can be selected in such a way that equivalent amounts of Cry6Aa and Cry55Aa are produced, so that the toxin combination becomes more effective against M. incognita. These toxin combinations might be used for the management of root‐knot nematodes because of their synergism in toxicity.
Experimental procedures
Bacterial strains and culture conditions
The recombinant B. thuringiensis strains BMB0215, BMB0250 and BMB0224 (Guo et al., 2008) were used for preparation of Cry6Aa, Cry5Ba and Cry55Aa respectively. All B. thuringiensis strains were cultured in ICPM medium (Guo et al., 2008) at 28°C with erythromycin (Sigma, 25 µg ml−1).
Crystal protein purification and quantification
The recombinant B. thuringiensis strains were cultured in ICPM medium (Guo et al., 2008) with 25 µg ml−1 erythromycin at 28°C, with shaking at 220 r.p.m. Cry6Aa, Cry5Ba and Cry55Aa proteins were then purified according to the method described by Griffitts et al., (2001). Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS‐PAGE) was used to examine the protein samples. The concentration of purified proteins was determined using the method of Bradford (1976), with bovine serum albumin (BSA) as the standard sample.
Rearing of M. incognita and bioassays
Meloidogyne incognita eggs were harvested from the root knots of infected tomato plants, and the M. incognita J2 were reared at 18–25°C and used in the bioassay. The bioassay procedure and evaluation of LC50 were undertaken according to the method described by Guo and colleagues (2008). The LC50 was determined using probit analysis (SAS 8.0). The expected LC50 value and synergistic factor was calculated using the formula of Tabashnik (1992), as follows:
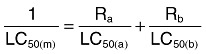 |
Where Ra and Rb are the proportions of toxin A and toxin B protein used in the final mixture; LC50(a) and LC50(b) are the LC50 values for each individual toxin; and LC50(m) is the expected theoretical LC50 value given by the harmonic mean of the intrinsic LC50 values of each component weighted by the ratio used in the mixture. The synergistic factor was calculated by dividing the expected toxicity [LC50(m)] by the observed toxicity of the mixture in bioassays. Synergistic factor values of more than 1 indicate synergism.
Phylogenetic tree and structure predictions
Amino acid sequence alignments and phylogenetic trees were produced using MEGA 4.1 (http://www.megasoftware.net/). Analysis of primary and secondary protein structure predictions was made using PHD (http://www.predictprotein.org/). The conserved blocks analysis was conducted using the methods reported by Schnepf and colleagues (1998).
Ligand blot analysis
Meloidogyne incognita J2 were harvested and frozen in liquid nitrogen. Total proteins were extracted by grinding and transferred onto a nitrocellulose membrane (Millipore, MA, USA). After blocking with 3% BSA in phosphate buffered saline (PBS)‐T buffer (0.1% Tween 20 in PBS, pH 7.4), nitrocellulose membranes were bathed in 5 ng ml−1 toxin for 2 h at room temperature. After washing three times with PBS‐T, membranes were bathed in respective anti‐Cry antibody for 2 h at 25°C and after washing three times with PBS‐T, the binding assay results were visualized with streptavidin–horseradish peroxide followed by SuperSignal chemiluminescent substrate (Pierce, FL, USA) as described by the manufacturers.
Acknowledgments
This study was supported by the National High Technology Research and Development Program (863) of China (2011AA10A203), the National Basic Research Program (973) of China (2009CB118902), the Genetically Modified Organisms Breeding Major Projects of China (2009ZX08009‐032B), and the China 948 Program of Ministry of Agriculture (2011‐G25) and Ministry of Forestry (2006–4‐41).
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Fig. S1. Evolutionary tree of crystal proteins documented against nematode. The phylogenetic trees were produced by the MEGA 4.1 software (http://www.megasoftware.net/).
Fig. S2. The conserved blocks comparison of crystal proteins documented against nematode. Analysis of primary and secondary protein structure predictions was made by the PHD software (http://www.predictprotein.org/). The conserved blocks analysis were conducted as the methods reported by Schnepf et al. (Schnepf et al., 1998).
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Bird D.M., Williamson V.M., Abad P., McCarter J., Danchin E.G., Castagnone‐Sereno P., Opperman C.H. The genomes of root‐knot nematodes. Annu Rev Phytopathol. 2009;47:333–351. doi: 10.1146/annurev-phyto-080508-081839. [DOI] [PubMed] [Google Scholar]
- Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein‐dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Bravo A., Gill S.S., Soberon M. Mode of action of Bacillus thuringiensis Cry and Cyt toxins and their potential for insect control. Toxicon. 2007;49:423–435. doi: 10.1016/j.toxicon.2006.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang S., Wang L., Guo W., Zhang X., Peng D., Luo C. Bacillus thuringiensis Bel protein enhances the toxicity of Cry1Ac protein to Helicoverpa armigera larvae by degrading insect intestinal mucin. Appl Environ Microbiol. 2009;75:5237–5243. doi: 10.1128/AEM.00532-09. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Frankenhuyzen K. Insecticidal activity of Bacillus thuringiensis crystal proteins. J Invertebr Pathol. 2009;101:1–16. doi: 10.1016/j.jip.2009.02.009. [DOI] [PubMed] [Google Scholar]
- Griffitts J.S., Haslam S.M., Yang T., Garczynski S.F., Mulloy B., Morris H. Glycolipids as Receptors for Bacillus thuringiensis Crystal toxin. Science. 2001;307:922–925. doi: 10.1126/science.1104444. et al. [DOI] [PubMed] [Google Scholar]
- Guo S., Liu M., Peng D., Ji S., Wang P., Yu Z., Sun M. New strategy for isolating novel nematicidal crystal protein genes from Bacillus thuringiensis strain YBT‐1518. Appl Environ Microbiol. 2008;74:6997–7001. doi: 10.1128/AEM.01346-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Chandra A., Pandey K.C. Bacillus thuringiensis (Bt) transgenic crop: an environment friendly insect‐pest management strategy. J Environ Biol. 2008;29:641–653. [PubMed] [Google Scholar]
- Li X.Q., Wei J.Z., Tan A., Aroian R.V. Resistance to root‐knot nematode in tomato roots expressing a nematicidal Bacillus thuringiensis crystal protein. Plant Biotechnol J. 2007;5:455–464. doi: 10.1111/j.1467-7652.2007.00257.x. [DOI] [PubMed] [Google Scholar]
- Li X.Q., Tan A., Voegtline M., Bekele S., Chen C.S., Aroian R.V. Expression of Cry5B protein from Bacillus thuringiensis in plant roots confers resistance to root‐knot nematode. Biol Control. 2008;47:97–102. [Google Scholar]
- Ohba M., Mizuki E., Uemori A. Parasporin, a new anticancer protein group from Bacillus thuringiensis. Anticancer Res. 2009;29:427–433. [PubMed] [Google Scholar]
- Perez C., Fernandez L.E., Sun J., Folch J.L., Gill S.S., Soberon M., Bravo A. Bacillus thuringiensis subsp. israelensis Cyt1Aa synergizes Cry11Aa toxin by functioning as a membrane‐bound receptor. Proc Natl Acad Sci USA. 2005;102:18303–18308. doi: 10.1073/pnas.0505494102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh J.Y., Choi J.Y., Li M.S., Jin B.R., Je Y.H. Bacillus thuringiensis as a specific, safe, and effective tool for insect pest control. J Microbiol Biotechnol. 2007;17:547–559. [PubMed] [Google Scholar]
- Rosas‐Garcia N.M. Biopesticide production from Bacillus thuringiensis: an environmentally friendly alternative. Recent Pat Biotechnol. 2009;3:28–36. doi: 10.2174/187220809787172632. [DOI] [PubMed] [Google Scholar]
- Sampson M.N., Gooday G.W. Involvement of chitinases of Bacillus thuringiensis during pathogenesis in insects. Microbiology. 1998;144:2189–2194. doi: 10.1099/00221287-144-8-2189. [DOI] [PubMed] [Google Scholar]
- Schnepf E., Crickmore N., Van Rie J., Lereclus D., Baum J., Feitelson J. Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol Mol Biol Rev. 1998;62:775–806. doi: 10.1128/mmbr.62.3.775-806.1998. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu C., Zhang J. Current patents related to Bacillus thuringiensis insecticidal crystal proteins. Recent Pat DNA Gene Seq. 2009;3:26–28. doi: 10.2174/187221509787236147. [DOI] [PubMed] [Google Scholar]
- Tabashnik B.E. Evaluation of synergism among Bacillus thuringiensis toxins. Appl Environ Microbiol. 1992;58:3343–3346. doi: 10.1128/aem.58.10.3343-3346.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trudgill D.L., Blok V.C. Apomictic, polyphagous root‐knot nematodes: exceptionally successful and damaging biotrophic root pathogens. Annu Rev Phytopathol. 2001;39:53–77. doi: 10.1146/annurev.phyto.39.1.53. [DOI] [PubMed] [Google Scholar]
- Wei J.Z., Hale K., Carta L., Platzer E., Wong C., Fang S.C., Aroian R.V. Bacillus thuringiensis crystal proteins that target nematodes. Proc Natl Acad Sci USA. 2003;100:2760–2765. doi: 10.1073/pnas.0538072100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z., Bai P., Ye W., Zhang F., Ruan L., Sun M. A novel negative regulatory factor for nematicidal Cry protein gene expression in Bacillus thuringiensis. J Microbiol Biotechnol. 2008;18:1033–1039. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Evolutionary tree of crystal proteins documented against nematode. The phylogenetic trees were produced by the MEGA 4.1 software (http://www.megasoftware.net/).
Fig. S2. The conserved blocks comparison of crystal proteins documented against nematode. Analysis of primary and secondary protein structure predictions was made by the PHD software (http://www.predictprotein.org/). The conserved blocks analysis were conducted as the methods reported by Schnepf et al. (Schnepf et al., 1998).


