Figure 4.
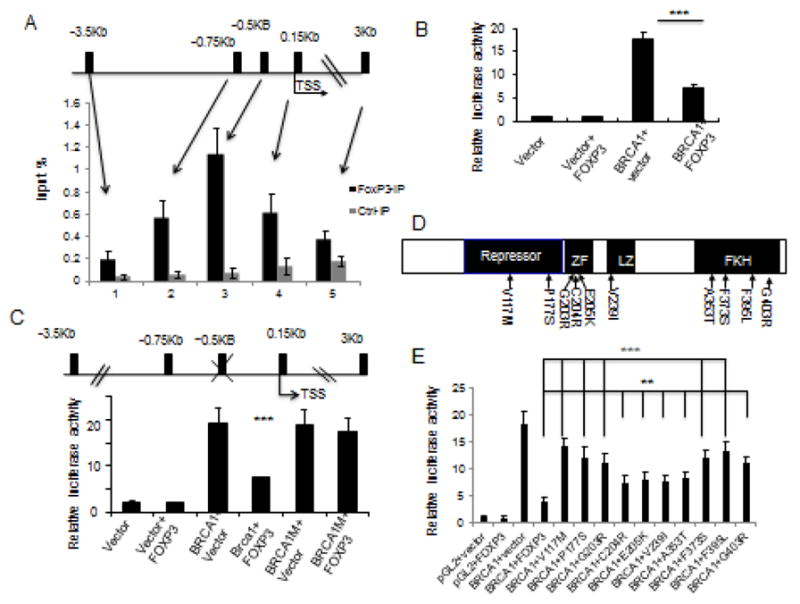
Identification of a functional FOXP3 binding site in the BRCA1 gene. A. The top panel shows a diagram of the potential forkhead binding motif in BRCA1 promoter, as marked black bars, while the lower panel shows that the % of input DNA precipitated by anti-FOXP3 or control IgG. Data shown are means+/−S.D. of % input DNA, from three independent experiments. TSS: transcriptional start site. B. Relative luciferase activity assay of BRCA1 promoter. 3.5 kB DNA upstream of BRCA1 TSS was cloned into luciferase reporter vector pGL2 transfected into 293 cells. Luciferase activity was assayed at 48 hours after transfection. Data shown are means+/−S.D. of three independent experiments. C. Identification of FOXP3 responsive element by mutational analysis. The FKH at −0.5 kB of BRCA1 TSS (GTCAACA) was mutated to GTCGGCA. The constructs used are diagrammed on the upper panel, while the transcriptional activity in the presence or absence of FOXP3 is presented in the lower panel. Data shown are means+/−S.D. of three independent analyses. D. Diagram depicting the position of somatic FOXP3 mutants from breast cancer samples used in this study. E. FOXP3 mutations abrogated its repression of BRCA1 promoter. Somatic mutations of FOXP3 from human breast and prostate cancer samples were tested for their repression of BRCA1 promoter using a luciferase reporter assay. Data shown are means+/−S.D. of triplicate sample analyses. **P<0.01, ***P<0.001.
