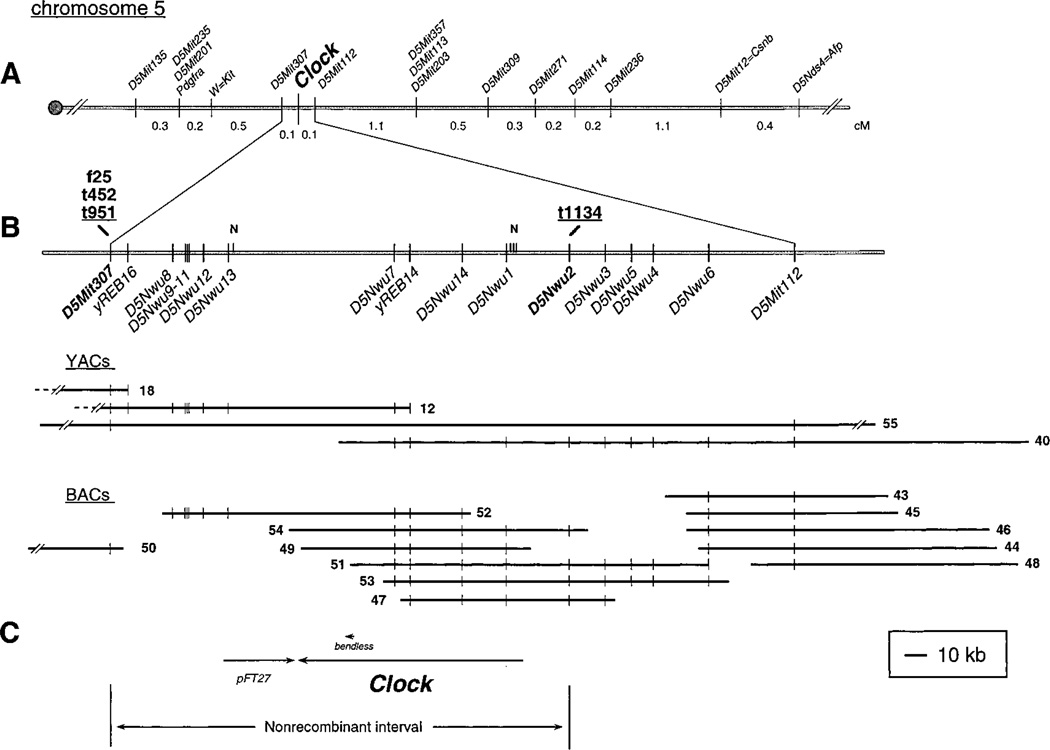
Figure 1. Genetic, Physical, and Transcription Unit Mapping of the Clock Locus.
(A) Genetic map of mouse chromosome 5, showing the location of Clock on the midportion of this chromosome, 0.7 cM distal of Kit, flanked by D5Mit307 and D5Mit112.
(B) Physical and high resolution genetic mapping of the Clock locus. The SSLPs and STSs in the D5Mit307–D5Mit112 interval are shown on the upper bar. The maximum nonrecombinant interval is defined by the SSLPs D5Mit307 and D5Nwu2. The animals with recombinations between these SSLPs and Clock are noted. Also shown are restriction sites for NotI (depicted with an [N]), which identify two CpG islands associated with the 5′ ends of Clock and pFT27. YAC and BAC contigs of the Clock region are shown below. Only four clones from the 32 clone YAC contig are shown. YACs 18 and 12 are chimeric clones and appear to be identical to YACs B16.S5.RE.C12 and B14.S4.RH.C8 in Brunkow et al. (1995); the part of each clone not mapping to this region of chromosome 5 is represented by a dashed line. The YACs were isolated from a library constructed by Kusumi et al. (1993), except for clone 55, which came from a library constructed by Larin et al. (1991). The BAC clones were isolated from a library constructed in Dr. Melvin Simon’s laboratory.
(C) Candidate genes mapping to the nonrecombinant interval. Shown are their relative locations and transcriptional orientations.