Abstract
Living cells are exquisitely responsive to mechanical cues, yet how cells produce and detect mechanical force remains poorly understood due to a lack of methods that visualize cell-generated forces at the molecular scale. Here we describe Förster resonance energy transfer (FRET)-based molecular tension sensors that allow us to directly visualize cell-generated forces with single-molecule sensitivity. We apply these sensors to determine the distribution of forces generated by individual integrins, a class of cell adhesion molecules with prominent roles throughout cell and developmental biology. We observe strikingly complex distributions of tensions within individual focal adhesions. FRET values measured for single probe molecules suggest that relatively modest tensions at the molecular level are sufficient to drive robust cellular adhesion.
Keywords: mechanobiology, integrin, focal adhesion, single molecule, tension, mechanotransduction, traction force microscopy
Disruption in the ability of cells to sense the mechanical properties of their surroundings represents a hallmark of many diseases, including muscular dystrophy, arteriosclerosis, cardiomyopathies, and cancer.1–3 Although cells have numerous mechanisms for detecting mechanical inputs, one of the most prominent is through integrins, heterodimeric transmembrane proteins that cluster into micron-sized assemblies termed focal adhesions (FAs).1, 4 FAs link the cell cytoskeleton to the surrounding extracellular matrix (ECM), and both transmit and respond to mechanical force.1, 5–7 Force transmission through integrins is essential for cell migration and adhesion, while force sensing at FAs regulates numerous cellular processes including proliferation and differentiation.8–10 The mechanisms by which integrins and their associated proteins both transmit and sense mechanical tension is therefore the subject of intense interest.
Force sensing at FAs has been proposed to occur at several levels of molecular complexity. Recent evidence suggests that mechanical load increases the affinity of α5β1 integrin for fibronectin, a canonical ECM component.11 This ‘catch bond’ model provides an appealing mechanism for increasing FA size and strength in response to mechanical load. Actomyosin-generated tension is likewise proposed to stretch FA components such as talin and p130Cas, exposing binding and phosphorylation sites that in turn recruit additional regulatory and cytoskeletal elements that reinforce the FA.7, 12, 13 Conversely, force-sensing and transmission may arise as a collective property of the FA. The proteins that link integrins to the actin cytoskeleton are known to be in rapid equilibrium, which allows the actin cytoskeleton to flow past FAs while still transmitting force. Relative slippage between actin and FA components has thus been proposed to regulate force transmission by functioning analogously to an automotive clutch.6, 14, 15
Although a great deal of evidence suggests that FAs undergo assembly, disassembly, and movement in a force-dependent manner, the underlying mechanisms remain poorly understood at the molecular level.1, 2, 5, 9 Reasonable estimates for the forces exerted at single integrins range over an order of magnitude, from 2–40 pN.16–20 It is likewise not known if all of the integrins within a single FA bear approximately the same tension, or instead experience a broad distribution of forces. Both the magnitude and spatiotemporal variations in the tension per integrin molecule have important consequences for mechanotransduction. A narrow range of molecular forces would argue for mechanical homeostasis at the level of individual integrins, while a broader distribution of forces would suggest that mechanosensing reflects a collective property of the FA as a whole. Differentiating between these and other potential models for mechanotransduction requires the measurement of the spatial distribution of tensions within FAs, and moreover the distribution of forces transmitted by individual integrins.
Current techniques for measuring cellular traction forces include deformable polymer substrates10, 21 and micropost arrays.22, 23 These techniques utilize the known material properties of the underlying substrates to calculate the magnitude and direction of stresses exerted by the cell based on the observed deformation of the substrate. However, both techniques report the local force averaged across many integrins, restricting the spatial resolution to approximately 1 micron.10 In addition, both approaches necessarily require that the substrate deform in order to observe cell-generated traction, introducing tradeoffs between the sensitivity of force detection and the accessible range of substrate stiffnesses. Recent work using fluorescence-based sensors complements traditional techniques, but lacks the single-molecule sensitivity necessary to dissect the molecular mechanisms governing force generation at FAs.16, 24–26
Here we report the development of FRET-based molecular tension sensors (MTSs) that report on the tensions experienced by single molecules in the presence of living cells (Figure 1a, b, S1). We developed these sensors by replacing the fluorescent proteins in a previously reported force probe16 with organic fluorophores with significantly higher brightness and photostablility.27 MTS probes allow the direct measurement of force maps with spatial resolution sufficient to distinguish variations in force within individual FAs. In addition, we used MTSs to explore the forces experienced by hundreds of individual integrins in live cells for the first time. Our work thus complements and extends recently reported approaches that use DNA-oligomer20 and gold-nanoparticle28 probes to measure cellular traction forces, but that lack the ability to track the forces experienced by single molecules in space and time.
Figure 1.
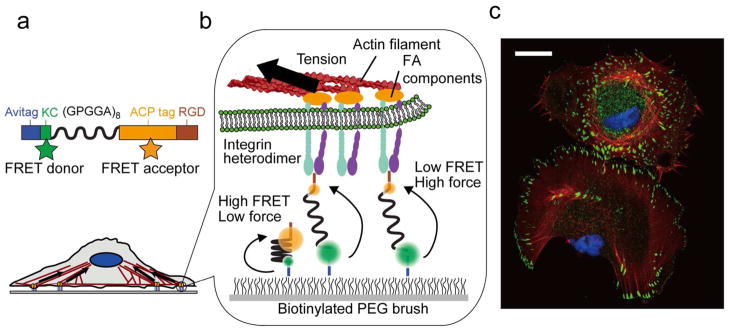
MTS overview. (a) Sensors are site-specifically labeled with biotin (Avitag), the FRET donor Alexa 546 (KC), and FRET acceptor CoA 647 (ACP tag) and present the RGD sequence from fibronectin (TVYAVTGRGDSPASSAA). The (GPGGA)8 sequence acts as an entropic spring that is stretched upon application of force. (b) Sensor molecules are attached to a coverslip via biotin and Neutravidin; the biotinylated PEG brush prevents nonspecific cell and sensor attachment. Integrin heterodimers attach to the RGD domain and apply load generated by the cell cytoskeleton. (c) Immunofluorescence image of fixed human foreskin fibroblast cells seeded on a MTS-functionalized surface; note the prominent actin stress fibers and FAs (blue: nucleus; red: actin; green: paxillin). Scale bar: 25 μm.
Human foreskin fibroblasts (HFFs) spread and develop stress fibers and robust FAs when seeded on glass surfaces functionalized with the MTS (Figure 1c), similarly to when they are seeded on collagen-coated surfaces (Figure S2). Our measurements employed an MTS density between 300 and 2000 molecules μm−2 (1 molecule per 20 nm × 20 nm area), an areal density known to support robust adhesion (Supporting Information, Figure S3).4, 29, 30 Cells do not adhere either to passivated surfaces or to surfaces functionalized with sensor molecules lacking the RGD sequence (Figure S2). These observations demonstrate that adhesion is mediated specifically by the RGD domain.
Preparing the coverslip surface with a high density of fluorophore-labeled MTS molecules results in a continuous fluorescent field. FRET images and time-lapse videos of multiple HFFs seeded on coverslips prepared in this way reveal regions of low FRET primarily near the cell periphery and in distinct patterns resembling FAs (Figure 2a, b, S4, S5, Supplementary Video S1). These areas coincide with paxillin recruitment, a canonical FA marker,31 showing that force generation is largely localized to FAs. However, closer inspection reveals that local regions of low FRET within FAs do not necessarily correspond to areas of maximum paxillin recruitment (Figure 2c, d).
Figure 2.
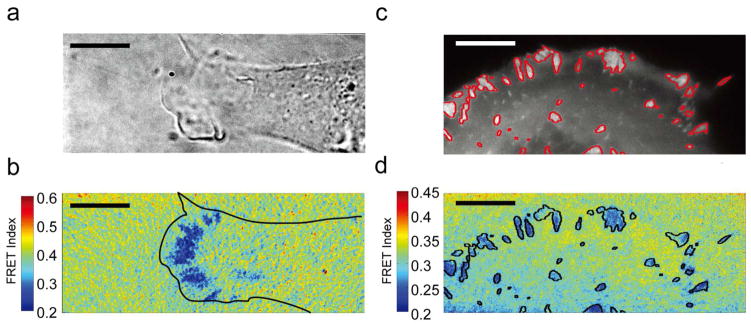
Ensemble FRET maps reveal regions of low FRET that colocalize with FAs. (a) Brightfield image of a spreading HFF on a MTS-functionalized surface. (b) Corresponding FRET map showing areas of high force localized at the cell periphery. Color bar represents FRET index (blue: low FRET/high force; red: high FRET/low force). (c) Live-cell image of an HFF expressing eGFP-paxillin with adhesions outlined in red. (d) Low FRET colocalizes with paxillin fluorescence, but paxillin levels do not strictly correlate with local force production. Scale bar: 10 μm.
Measurements performed with a control probe that is inert to force support the likelihood that tension propagated through the force-sensing module is responsible for FRET changes (Figure S6, S7). In this construct the biotinylation site is C-terminal to the RGD sequence. Thus, cellular traction force should be propagated directly to the coverslip, bypassing the fluorophore-flanked (GPGGA)8 spring (Figure S6). Indeed, we did not observe FRET changes with this sensor molecule even in areas of robust paxillin recruitment (Figure S7). The FRET signal observed with the MTS is thus unlikely to reflect changes in fluorophore orientation or local environment caused by integrin binding. Additionally, the MTS FRET response is dissipated within 5 minutes when cells are treated with cytochalasin D, a cytoskeletal inhibitor that prevents actin polymerization (Figure S8). Together, these observations indicate that MTS molecules report cytoskeletally generated tension transmitted via integrins.
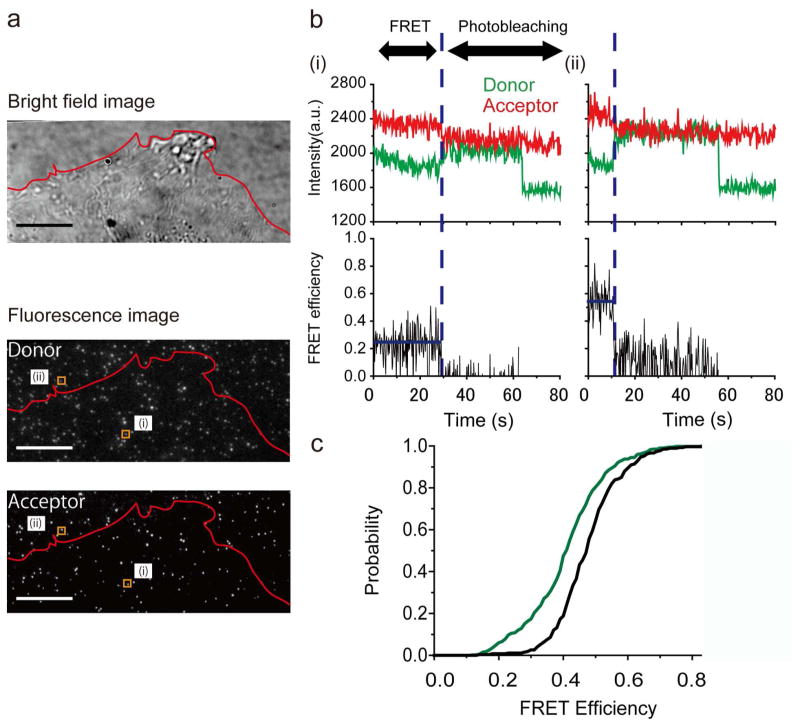
In order to observe FRET events at individual molecules, we diluted fluorophore-labeled MTS molecules approximately ~1:1,000 with the unlabeled MTS (see Materials and Methods, Supporting Information). This preserved the RGD surface density necessary for cell adhesion while providing sufficient separation between fluorophore-labeled sensors to allow them to be imaged individually (~1 labeled MTS per 2 μm2). Individual labeled sensor molecules are clearly visible underneath live cells (Figure 3a). We analyzed only molecules in which we observed sequential, single-step photobleaching of the acceptor and donor fluorophores. This photobleaching sequence confirms that the molecules analyzed were individual MTS probes singly-labeled with both fluorophores (Figure 3b, S9).27
Figure 3.
Force exerted by single integrins. (a) HFFs spreading on a MTS-coated coverslip visualized in brightfield; individual molecules are clearly visible in the donor and acceptor channels. Scale bar: 10 μm. The outlined regions (i) and (ii) correspond to molecules exhibiting high and low FRET, and residing outside the cell boundary and underneath the cell, respectively. (b) Raw single-molecule FRET traces from molecules (i) and (ii). Two-step photobleaching indicates the presence of a doubly-labeled sensor. (c) Cumulative probability distributions of FRET values measured for individual MTS molecules in the presence (green; 398 molecules, 13 cells) and absence of cells (black; 266 molecules). The distributions are statistically distinct (2 sample Kolmogorov-Smirnov test, p-value: 1.7×10−12).
MTS molecules outside of the area covered by the cell show FRET efficiencies of ~0.5, consistent with single-molecule FRET measurements made in the absence of cells (Figures 3c, S10). While some MTS molecules underneath the cell also show FRET values consistent with zero applied force, a subpopulation of molecules show lower FRET efficiencies indicating that they are under tension (Figures 3c, S10). In contrast, FRET values measured in the presence and absence of cells using MTS molecules lacking the RGD domain are statistically indistinguishable with a p-value > 0.05 (Figures S11, S12). This observation indicates that changes in FRET observed with the full-length MTS result from cellular traction forces transmitted specifically via the RGD domain.
Tensions that stretch the MTS appreciably beyond the Förster radius would be anticipated to result in undetectably low FRET. To test for this possibility, we first identified colocalized donor and acceptor fluorophores via simultaneous excitation at 532 and 635 nm, and then examined these pairs to determine the fraction that showed < 5% FRET. Approximately 7% of coincident donor and acceptor fluorophores show undetectably low FRET. However, these “zero FRET” pairs are observed with approximately equal probability both underneath cells and in regions of the coverslip where cells are not attached. It is therefore likely that many of these instances reflect colocalization of two MTS molecules, each containing either a working FRET donor or acceptor, or MTS molecules in which the FRET acceptor photobleaches during the first few frames of the FRET measurement (Supporting Information). These observations suggest that the force exerted on individual probes falls within the detection limits of the MTS in the large majority of cases. However, we cannot exclude the possibility that a small population of MTSs may experience forces outside the detectable range.
Integrins are known to play a central role in both cell motility and mechanotransduction, but the force generated at individual integrin complexes is not known. Previous work reports a FRET vs. force calibration curve for the (GPGGA)8 molecular spring.16 In principle, this provides a means to estimate forces experienced by individual molecules in our assay. Based on this calibration, we observe that most MTS molecules are not under tension, while a subpopulation experiences tensions of approximately 1 to 5 pN (Figure S14). This distribution is reasonable given that the majority of measured MTS molecules likely fall outside regions of force production. We note that although the protein sequence and fluorophores used in the previous calibration are similar to those in our experiment, they are not identical. While force estimation is relatively insensitive to variations in Förster radius (Figure S13), differences in protein sequence, for example the presence of the ACP labeling domain, may affect the force calibration.
Although the absolute forces derived from our measurement are necessarily approximate, they are informative in the context of previous measurements. Optical trap and AFM measurements of integrin contact rupture forces are up to an order of magnitude larger than the forces we calculate.18, 19, 32, 33 A recently reported DNA-based sensor designed to detect maximal forces transmitted through integrins likewise suggests peak forces of 30–40 pN.20 These observations may indicate that the maximal load supported by integrins is considerably more than is generated at equilibrium. The forces we calculate match remarkably well with the single-pN tensions inferred from ensemble FRET measurements of a genetically-encoded vinculin tension sensor,16 and from a gold-nanoparticle based sensor that reports on tensions experienced by cyclic RGD ligands.28 Our data are also consistent with the ~2 pN slip bond inferred to exist between talin and actin.17 The single-pN forces we measure agree with traction force measurements: a reasonable estimated density of ~500 integrins per μm2 and 2 pN per integrin yields a total traction force of 2 nN for a FA with 2 μm2 surface area, corresponding to a traction stress of ~1 kPa.4, 10, 29, 34 We note that the RGD sequence used in the MTS contains only a portion of the integrin binding site in fibronectin. Previous work suggests that integrins adopt multiple distinct conformations when binding to fibronectin, and that high-affinity binding and activation may require engagement with a secondary binding site (PHSRN sequence).35–38 More broadly, differences in the integrin heterodimer, ligand, and activation state39 could result in higher (or lower) tensions than are observed here.
High-resolution traction force microscopy measurements indicate that traction forces localize towards the distal FA tip in mouse embryonic fibroblasts.40 Our ensemble measurements are generally consistent with this result. However, the additional detail provided by our measurements reveals that the distribution of forces within individual FAs is strikingly complex (Figure 2d). In particular, we observe that paxillin recruitment, a standard FA marker, does not necessarily correlate with regions of high force production.
Although subject to important provisos, FRET measurements from individual MTS molecules provide useful estimates of the forces transmitted by single integrins. The sub-10 pN forces we observe suggest that relatively weak interactions at the molecular scale are sufficient to drive robust cell adhesion. Cell-ECM adhesion, and by extension mechanotransduction, may thus reflect the collective contribution of numerous weak interactions. However, integrin complexes are capable of resisting much higher loads than we observe.16–20, 32, 33 Whether mechanotransduction results from weak, steady-state interactions or instead from transient but higher forces provides an important target for future research.
Numerous classes of adhesion proteins link cells to each other and the ECM in addition to integrins. How cells may sense mechanical input at these sites is essentially unknown. The MTS design is readily generalizable to other cellular adhesions and is thus potentially applicable to studying force transmission at cadherin complexes25 and the many other adhesive interactions that link the cell to its surroundings. MTS measurements are thus highly suited to the discovery and characterization of mechanosensory pathways that are at present largely unexplored.
Supplementary Material
Acknowledgments
We thank the members of the A.R.D. laboratory for insightful discussions, Jongmin Sung (Stanford University) for help with the TIRF setup, and Tristan Ursell (Stanford University) for custom MATLAB image analysis routines. This work is supported by a Stanford Bio-X IIP award (ARD), the National Science Foundation (NSF) under Emerging Frontiers in Research and Innovation (EFRI) Grant 1136790 (ARD), National Institute of Health (NIH) New Innovator Award 1DP2OD007078 (ARD), Stanford Cardiovascular Institute Seed Grant (ARD), Burroughs-Wellcome Career Award at the Scientific Interface (ARD), NSF Graduate Research Fellowship 1000121811 (AHM), and a Stanford Graduate Fellowship (ASA). We also acknowledge the support of the Stanford Neuroscience Microscopy Service (NIH NS069375) under Andrew Olson for confocal imaging and technical advice.
Funding Sources
This work is supported by a Stanford Bio-X IIP award (ARD), the National Science Foundation (NSF) under Emerging Frontiers in Research and Innovation (EFRI) Grant 1136790 (ARD), National Institute of Health (NIH) New Innovator Award 1DP2OD007078 (ARD), Stanford Cardiovascular Institute Seed Grant (ARD), Burroughs-Wellcome Career Award at the Scientific Interface (ARD), NSF Graduate Research Fellowship 1000121811 (AHM), and a Stanford Graduate Fellowship (ASA).
Abbreviations
- FA
focal adhesion
- ECM
extracellular matrix
- FRET
Förster resonance energy transfer
- MTS
molecular tension sensor
- HFF
human foreskin fibroblast
- RGD
arginine-glycine-aspartic acid
- ACP
acyl carrier protein
- PEG
polyethylene glycol
- eGFP
enhanced green fluorescent protein
- AFM
atomic force microscopy
Footnotes
Author Contributions
The manuscript was written through contributions from all of the authors. All authors have given approval to the final version of the manuscript.
Detailed materials and methods, supplementary figures, and supplementary videos are available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Hoffman BD, Grashoff C, Schwartz MA. Nature. 2011;475:316–323. doi: 10.1038/nature10316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jaalouk DE, Lammerding J. Nat Rev Mol Cell Biol. 2009;10:63–73. doi: 10.1038/nrm2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Butcher DT, Alliston T, Weaver VM. Nat Rev Cancer. 2009;9:108–122. doi: 10.1038/nrc2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu CH, Law JB, Suryana M, Low HY, Sheetz MP. Proc Natl Acad Sci U S A. 2011;108(51):20585–20590. doi: 10.1073/pnas.1109485108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aratyn-Schaus Y, Gardel ML. Curr Biol. 2010;20:1145–1153. doi: 10.1016/j.cub.2010.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gardel ML, Sabass B, Ji L, Danuser G, Schwarz US, Waterman CM. J Cell Biol. 2008;183:999–1005. doi: 10.1083/jcb.200810060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wolfenson H, Bershadsky A, Henis YI, Geiger B. J Cell Sci. 2011;124:1425–1432. doi: 10.1242/jcs.077388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, Parsons JT, Horwitz AR. Science. 2003;302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 9.Palecek SP, Loftus JC, Ginsberg MH, Lauffenburger DA, Horwitz AF. Nature. 1997;385:537–540. doi: 10.1038/385537a0. [DOI] [PubMed] [Google Scholar]
- 10.Sabass B, Gardel ML, Waterman CM, Schwarz US. Biophys J. 2008;94:207–220. doi: 10.1529/biophysj.107.113670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Friedland JC, Lee MH, Boettiger D. Science. 2009;323:642–644. doi: 10.1126/science.1168441. [DOI] [PubMed] [Google Scholar]
- 12.Sawada Y, Tamada M, Dubin-Thaler BJ, Cherniavskaya O, Sakai R, Tanaka S, Sheetz MP. Cell. 2006;127:1015–1026. doi: 10.1016/j.cell.2006.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.del Rio A, Perez-Jimenez R, Liu R, Roca-Cusachs P, Fernandez JM, Sheetz MP. Science. 2009;323:638–641. doi: 10.1126/science.1162912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parsons JT, Horwitz AR, Schwartz MA. Nat Rev Mol Cell Biol. 2010;11:633–643. doi: 10.1038/nrm2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu K, Ji L, Applegate KT, Danuser G, Waterman-Storer CM. Science. 2007;315:111–115. doi: 10.1126/science.1135085. [DOI] [PubMed] [Google Scholar]
- 16.Grashoff C, Hoffman BD, Brenner MD, Zhou R, Parsons M, Yang MT, McLean MA, Sligar SG, Chen CS, Ha T, Schwartz MA. Nature. 2010;466:263–266. doi: 10.1038/nature09198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang G, Giannone G, Critchley DR, Fukumoto E, Sheetz MP. Nature. 2003;424:334–337. doi: 10.1038/nature01805. [DOI] [PubMed] [Google Scholar]
- 18.Thoumine O, Kocian P, Kottelat A, Meister JJ. Eur Biophys J. 2000;29:398–408. doi: 10.1007/s002490000087. [DOI] [PubMed] [Google Scholar]
- 19.Li F, Redick SD, Erickson HP, Moy VT. Biophys J. 2003;84:1252–1562. doi: 10.1016/S0006-3495(03)74940-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang X, Ha T. Science. 2013;340:991–994. doi: 10.1126/science.1231041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harris AK, Wild P, Stopak D. Science. 1980;208:177–179. doi: 10.1126/science.6987736. [DOI] [PubMed] [Google Scholar]
- 22.Tan JL, Tien J, Pirone DM, Gray DS, Bhadriraju K, Chen CS. Proc Natl Acad Sci U S A. 2003;100(4):1484–1489. doi: 10.1073/pnas.0235407100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.du Roure O, Saez A, Buguin A, Austin RH, Chavrier P, Silberzan P, Ladoux B. Proc Natl Acad Sci U S A. 2005;102(7):2390–2395. doi: 10.1073/pnas.0408482102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meng F, Suchyna TM, Sachs F. FEBS J. 2008;275:3072–3087. doi: 10.1111/j.1742-4658.2008.06461.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Borghi N, Sorokina M, Shcherbakova OG, Weis WI, Pruitt BL, Nelson WJ, Dunn AR. Proc Natl Acad Sci U S A. 2012;109(31):12568–12573. doi: 10.1073/pnas.1204390109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stabley DR, Jurchenko C, Marshall SS, Salaita KS. Nat Methods. 2012;9:64–67. doi: 10.1038/nmeth.1747. [DOI] [PubMed] [Google Scholar]
- 27.Roy R, Hohng S, Ha T. Nat Methods. 2008;5:507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Y, Yehl K, Narui Y, Salaita K. J Am Chem Soc. 2013;135(14):5320–5323. doi: 10.1021/ja401494e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cavalcanti-Adam EA, Volberg T, Micoulet A, Kessler H, Geiger B, Spatz JP. Biophys J. 2007;92:2964–2974. doi: 10.1529/biophysj.106.089730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang J, Grater SV, Corbellini F, Rinck S, Bock E, Kemkemer R, Kessler H, Ding J, Spatz JP. Nano Lett. 2009;9(3):1111–1116. doi: 10.1021/nl803548b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laukaitis CM, Webb DJ, Donais K, Horwitz AF. J Cell Biol. 2001;153(7):1427–1440. doi: 10.1083/jcb.153.7.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Litvinov RI, Shuman H, Bennett JS, Weisel JW. Proc Natl Acad Sci U S A. 2002;99(11):7426–7431. doi: 10.1073/pnas.112194999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun Z, Martinez-Lemus LA, Trache A, Trzeciakowski JP, Davis GE, Pohl U, Meininger GA. Am J Physiol: Heart Circ Physiol. 2005;289:H2526–H2535. doi: 10.1152/ajpheart.00658.2004. [DOI] [PubMed] [Google Scholar]
- 34.Shibata ACE, Fujiwara TK, Chen L, Suzuki KGN, Ishikawa Y, Nemoto YL, Miwa Y, Kalay Z, Chadda R, Naruse K, Kusumi A. Cytoskeleton. 2012;69:380–392. doi: 10.1002/cm.21032. [DOI] [PubMed] [Google Scholar]
- 35.Aota S, Nomizu M, Yamada KM. J Biol Chem. 1994;269:24756–24761. [PubMed] [Google Scholar]
- 36.Chen W, Lou J, Evans EA, Zhu C. J Cell Biol. 2012;199:497–512. doi: 10.1083/jcb.201201091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Askari JA, Buckley PA, Mould AP, Humphries MJ. J Cell Sci. 2009;122:165–170. doi: 10.1242/jcs.018556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Askari JA, Tynan CJ, Webb SED, Martin-Fernandez ML, Ballestrem C, Humphries MJ. J Cell Biol. 2010;188(6):891–903. doi: 10.1083/jcb.200907174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garcia AJ, Huber F, Boettiger D. J Biol Chem. 1998;273:10988–10993. doi: 10.1074/jbc.273.18.10988. [DOI] [PubMed] [Google Scholar]
- 40.Plotnikov SV, Pasapera AM, Sabass B, Waterman CM. Cell. 2012;151:1513–1527. doi: 10.1016/j.cell.2012.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.