Figure 2.
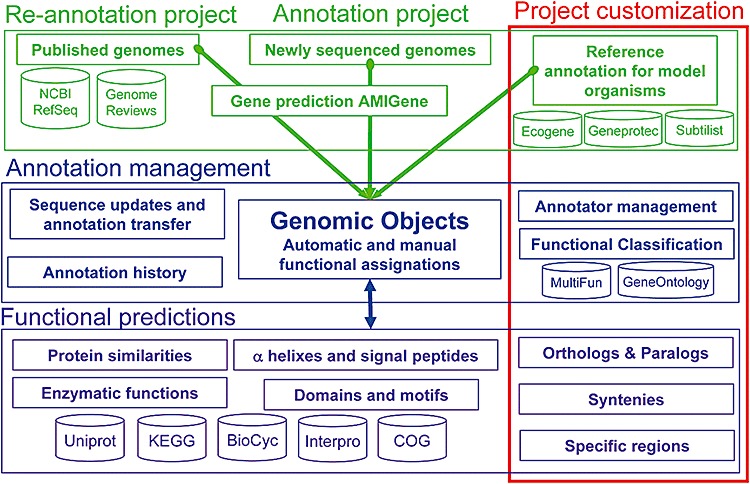
Simplified prokaryotic genome database (PkGDB) relational model composed of three main components: sequence and annotation data (in green), annotation management (in blue) and functional predictions (in purple). Sequences and annotations come from public databanks, sequencing centres and specialized databases focused on model organisms. For genomes of interest, a (re)‐annotation process is performed using AMIGene (Bocs et al., 2003) and leads to the creation of new ‘Genomic Objects’. Each ‘Genomic Object’ and associated functional prediction results are stored in the PkGDB. The database architecture supports integration of automatic and manual annotations, and management of a history of annotations and sequence updates. Reproduced from Vallenet and colleagues (2006).
