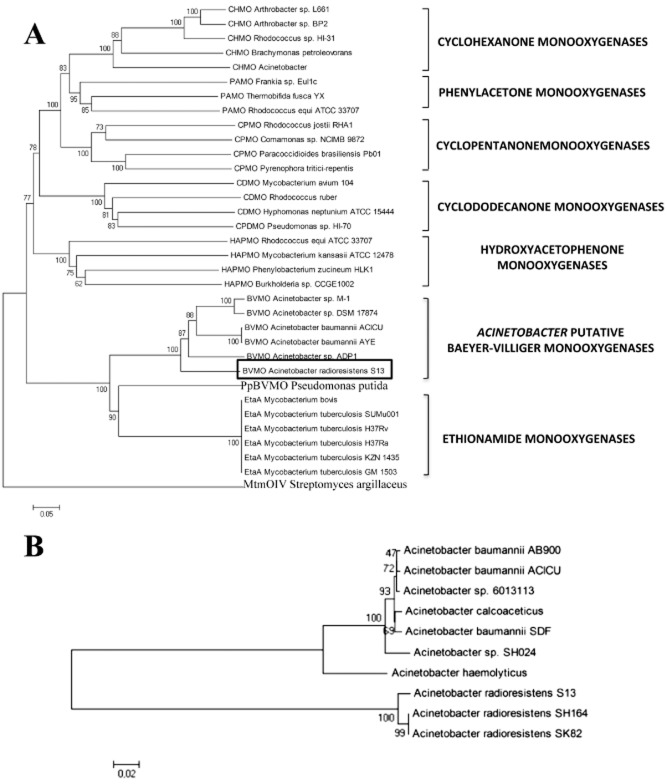
Figure 5.
Phylogenetic relationships within Baeyer‐Villiger monooxygenases (A) and alkane monooxygenases (B). Sequences of proteins with confirmed BVMO and alkane monooxygenase activity were aligned and the un‐rooted phylogenetic trees were calculated by using clustalw and neighbour‐joining method. The numbers at the nodes are the bootstrap confidence values obtained after 1000 replicates. The scale bar indicates distance in substitutions per nucleotide. The species and proteins are: (A) CHMO A rthrobacter sp. L661 = cyclohexanone monooxygenase (ABQ10653.1); CHMO A rthrobacter sp. BP2 = cyclohexanone monooxygenase (AA N37479.1); CHMO R hodococcus sp. HI‐31 = cyclohexanone monooxygenase (BAH56670); CHMO B rachymonas petroleovorans = cyclohexanone monooxygenase (AAR99068.1); CHMO A cinetobacter = cyclohexanone monooxygenase (AF282240_5); PAMO Frankia sp. Eu1c = phenylacetone monooxygenase (YP_004016782.1); PAMO T hermobifida fusca YX = phenylacetone monooxygenase (AAZ55526.1); PAMO R hodococcus equi ATCC33707 = phenylacetone monooxygenase (ZP_ 08156844.1); CPMO R hodococcus josti RHA1 = cyclopentanone monooxygenase (YP_703208.1); CPMO C omamonas sp. NCIMB 9872 = cyclopentanone monooxygenase (BAC22652.1); CPMO P aracoccidioides brasiliensis Pb01 = cyclopentanone monooxygenase (XP_002792362.1); CPMO P yrenophora treitici‐repentis = cyclopentanone monooxygenase (XP_001942142.1); CDMO M ycobacterium avium 104 = cyclododecanone monooxygenase (YP_884322.1); CDMO R hodococcus ruber = cyclododecanone monooxygenase (AAL14233.1); CDMO H yphomonas neptunium ATCC15444 = cyclododecanone monooxygenase (YP_760004.1); CPDMO P seudomonas sp. HI‐70 = cyclopentadecanone monooxygenase (BAE93346.1); HAPMO R hodococcus equi ATCC33707 = hydroxyacetophenone monooxygenase (ZP_08156846.1); HAPMO M ycobacterium kansasii ATCC33707 = hydroxyacetophenone monooxygenase (ZP_04749205.1); HAPMO P henylobacterium zucineum HLK1 = hydroxyacetophenone monooxygenase (YP_002130647.1); Burkholderia sp. CCGE 1002 = hydroxyacetophenone monooxygenase (ADG19710.1); BVMO A cinetobacter sp. M‐1 = alkane monooxygenase (ABQ18228.1); BVMO A cinetobacter sp. DSM 17874 = Baeyer‐Villiger monooxygenase (ABQ18224.1); BVMO A cinetobacter baumannii ACICU = Baeyer‐Villiger monooxygenase (YP_001847910.1); BVMO A cinetobacter baumannii ACICU = Baeyer‐Villiger monooxygenase (YP_001847910.1); BVMO A cinetobacter baumannii AYE = Baeyer‐Villiger monooxygenase (YP_001712414); BVMO A cinetobacter sp. ADP1 = Baeyer‐Villiger monooxygenase (YP_047698.1); BVMO A cinetobacter radioresistens S13 = Baeyer‐Villiger monooxygenase (GU145276.1); PpBVMO P seudomonas putida = monooxygenase of flavin‐binding family (AAN68413.1); EtaA M ycobacterium bovis BCG = ethionamide monooxygenase (YP_979996.1); EtaA M ycobacterium tuberculosis SUMu001 = ethionamide monooxygenase (ZP_07416557.1); EtaA M ycobacterium tuberculosis H37Rv = ethionamide monooxygenase (NP_218371.1); EtaA M ycobacterium tuberculosis H37Ra = ethionamide monooxygenase (YP_001285245.1); EtaA M ycobacterium tuberculosis KZN1435 = ethionamide monooxygenase (YP_003033907); EtaA M ycobacterium tuberculosis GM1503 = ethionamide monooxygenase (ZP_03534438.1), MtmOIV S treptomyces argillaceus = mithramycin monooxygenase (3FMW_A); (B) Acinetobacter baumannii AB900 = terminal alkane‐1‐monooxygenase (ZP_04661203.1); Acinetobacter baumannii ACICU = terminal alkane‐1‐monooxygenase (YP_001846325.1); Acinetobacter sp. 6013113 = alkane‐1‐monooxygenase (ZP_06781771.1); Acinetobacter calcoaceticus = alkane‐1‐monooxygenase (CAB51020.1); Acinetobacter baumannii SDF = terminal alkane‐1‐monooxygenase (YP_001707231.1); Acinetobacter radioresistens sp. SH164 = alkane hydroxylase B (ZP_06072466.1); Acinetobacter haemolyticus = alkane hydroxylase (AAS93604.4); Acinetobacter radioresistens S13 = alkane hydroxylase B (HQ68589); Acinetobacter sp. SH164 = alkane hydroxylase B (ZP_06072466.1); Acinetobacter radioresistens SK82 = alkane‐1‐monooxygenase (ZP_05361594.1).