Figure 4.
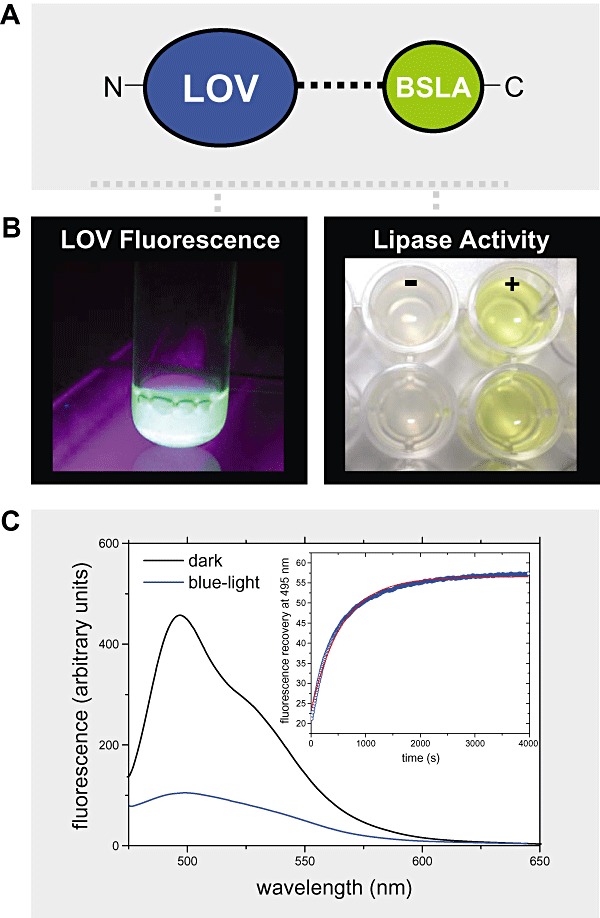
A and B. (A) Illustration of the fusion strategy: a bacterial LOV domain (here of the B. subtilis YtvA protein) was fused N‐terminally to the lipase A of B. subtilis (BSLA) resulting in the construct nLOV‐BSLA. The two protein parts are linked via a helical linker polypeptide naturally present in B. subtilis YtvA. The fusion construct can be readily expressed in soluble form and retains both chromophore binding capacity in the LOV domain as evidenced by flavin fluorescence (left part of B) and lipolytic activity of the lipase (right part of B). Here shown is the result of a standard lipase activity assay using p‐nitrophenylpalmitate (pNPP) as substrate (+). The corresponding negative control (−) contained no enzyme. The two rows of wells depict the results of a duplicate measurement. The lipase cleaves the ester‐bond of pNPP resulting in the formation of the yellow‐coloured p‐nitrophenylate anion indicative for enzymatic activity. C. Observed light sensitivity of nLOV‐BSLA: Flavin‐fluorescence was recorded in the proteins dark state (black line) as well as 30 s after blue‐light illumination (blue line). In the dark, the protein displays a bimodal fluorescence with maxima occurring at around 495 and 520 nm. Upon formation of the signalling state, this fluorescence is effectively quenched by the formation of the covalent flavin‐cysteinyl adduct. The inset shows the dark‐recovery time trace of the FMN fluorescence peak recorded at 495 nm.
