Figure 3.
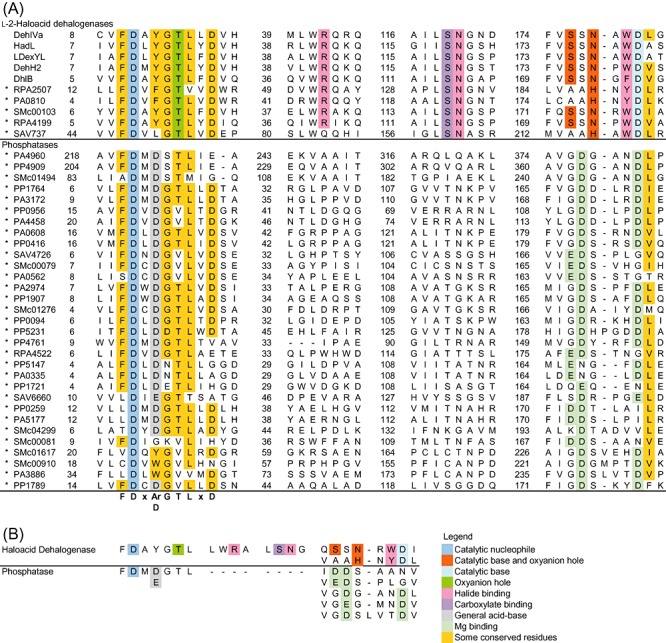
Sequence analyses of HAD targets in the initial screen. The conserved residues in both enzyme groups are presented. A. The sequence alignment of HAD targets identified in the initial screen (*). The overall sequence similarity is higher among the l‐2‐haloacid dehalogenases. The catalytic Asp nucleophile is located at the N‐terminal motif conserved in both enzyme groups. At position nucleophile + 2, the l‐2‐haloacid dehalogenases generally possess an aromatic residue while most phosphatases carry a second Asp. Near the C‐terminus, the dehalogenases and phosphatases have distinct sequence motifs containing catalytically important residues. The numbers mark the residue number for the residue to their right. B. The key catalytic motifs extracted from the sequence analysis. The conserved motif variations within each subfamily are also shown.
