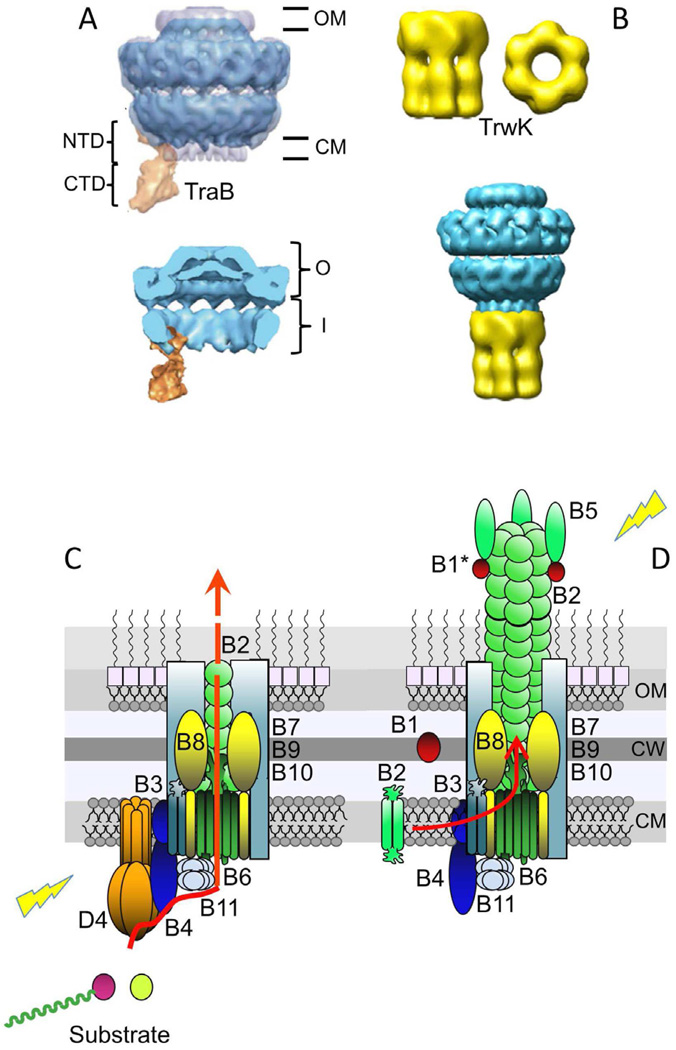
Fig. 2.
Gram-negative T4SS subassemblies and architectural models. (A) Upper: Structure of the pKM101-encoded core complex with the VirB4-like TraB homolog bound to the side. The core is shown in blue and TraB is in orange. The O and I layers are indicated as O and I, respectively. TraBN-terminal domain (NTD) and C-terminal domain (CTD) are shown. The negative stain core/TraB (in blue) is superimposed over the cryoelectron microscopy structure of the core complex alone (half-gray transparent). Lower: Section view of the core/TraB structure showing the core chamber, which is envisioned to house the translocation channel and pilus (Wallden et al., 2012). Images were published with permission. (B) Upper: Three dimensional reconstruction of VirB4-like TrwK of plasmid R388 from negative-stained images. Side and bottom views of the hexamer are shown. Lower: Model of interaction between the T4SS core complex and TrwKR388. The TrwKR388 hexameric structure was attached to the cytoplasmic side of the inner membrane region of the pKM101 core complex obtained from (Fronzes et al., 2009b). These images were reproduced with permission by (Pena et al., 2012). The figure is not to scale, dimensions of the core are ∼185 Å in width and length (Fronzes et al., 2009b), and those of the TrwKR388 hexamer are ∼132 Å and ∼165 Å in width and length, respectively (Pena et al., 2012). (C) Model depicting the architecture of the VirB/VirD4 translocation channel in the Gram-negative cell envelope, with the substrate translocation pathway depicted (red arrow). VirD4At and VirBAt channel subunits are shown (color code matches Fig. 1), with the shaded aqua rectangles corresponding to the VirB7/VirB9/VirB10 core complex. VirD4 T4CP is required for substrate transfer. Substrates include relaxase-T-strand (left) and effector protein (right). (D) Model of VirB/VirD4 conjugative pilus. Pilus biogenesis involves VirB4-mediated dislocation of membrane pilins and entry into the core chamber for polymerization. Pilus polymerization requires substantial conformational changes in the core complex. VirB1/VirB1*, but not VirD4 are required for pilus assembly. Lightning bolts: The T4SS apparatus is activated by substrate engagement (left) and target cell or bacteriophage binding to the extracellular pilus (right). Cytoplasmic membrane (CM), cell wall (CW), outer membrane (OM).