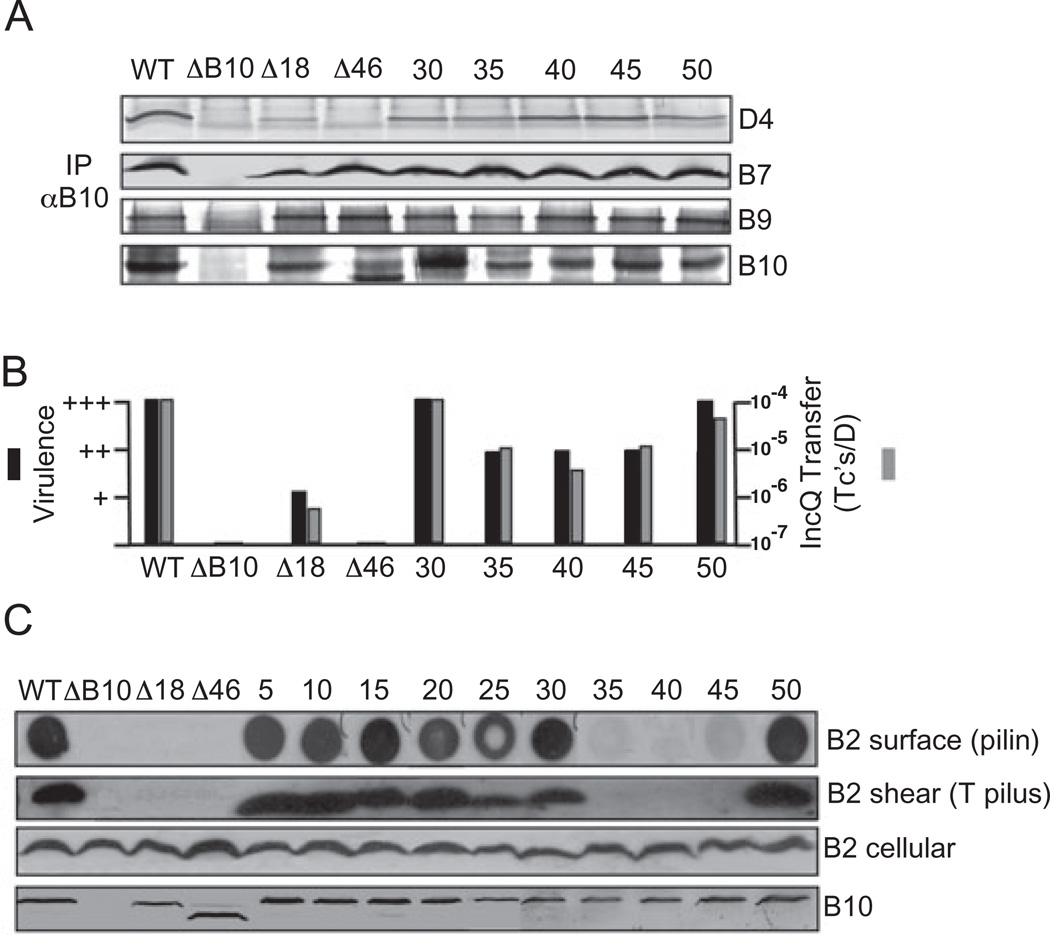
Fig. 2. Phenotypes of N-terminal insertion and deletion mutations.
A. Effects of mutations on VirB10 partner interactions. Material immunoprecipitated from detergent-solubilized cell extracts with anti-VirB10 antibodies (IP αB10) was analysed for the presence of VirD4, VirB7, VirB9 and VirB10 by immunoblot development with corresponding antibodies. Experiments were carried out under non-reducing conditions for co-precipitation of the VirB7–VirB9 dimer complex.
B. Effects of mutations on T-DNA transfer as monitored by virulence on wounded Kalanchoe leaves (black bars; - avirulent, +++ WT virulence) and transfer of the mobilizable IncQ plasmid pML122 to A. tumefaciens recipients (Grey bars; Tc’s/D, the number of transconjugants per donor cell).
C. Effects of mutations on T pilus production. Top-to-bottom: colony immunoblots developed with anti-VirB2 antibodies showing presence of surface-exposed pilin protein (B2 surface); extracellular shear fraction subjected to ultracentrifugation, gel electrophoresis and immunoblot development with anti-VirB2 antibodies revealing presence of high-molecular-weight T pilus (B2 shear); total cellular material subjected to gel electrophoresis and blot development with anti-VirB2 or anti-VirB10 antibodies revealing total cellular levels of the respective proteins. Strains: WT, wild-type Δ348; ΔB10, non-polar vIrB10 strain PC1010; Δ18, PC1010 producing VirB10Δ1–18; A46, PC1010 producing VirB10Δ1–46 fused to the VirB5 signal sequence for export of the C-terminal fragment to the periplasm; 5–50, PC1010 producing i2 (Ala-Cys) insertion mutations at the residue indicated. Strains producing mutant proteins with i2 insertions at residues 5, 10, 15, 20 and 25 displayed WT phenotypes with respect to co-immunoprecipitation (A) and substrate transfer (B) (data not shown).