Figure 3.
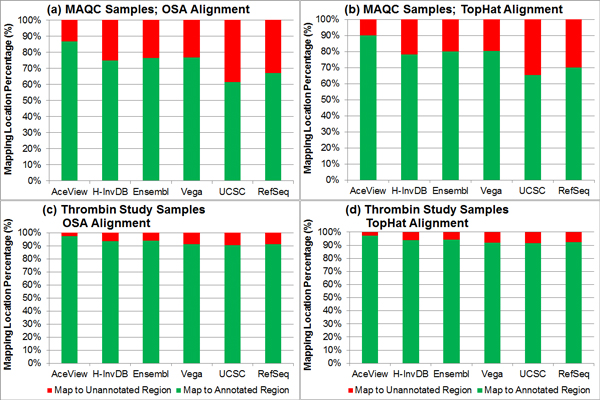
The percentage of reads or read pairs that map to the annotated and un-annotated genomic sequences. Sub-figures (a)-(d) represent different combinations of samples (top: MAQC samples with SRA accession number SRP000727; bottom: thrombin study samples with SRA accession number SRP008482) and spliced mappers (left: OSA; right: TopHat). The UCSC annotation usually has the lowest percentage of reads that mapped to the annotated genomic sequences, while the AceView annotation usually has the highest percentage. The same observation is applicable to all four combinations of samples and spliced mappers.
