Table 3. Sequence diversity and distribution of naturally occurring oxytocin-like nonapeptides.
| Peptide | Sequence‡ | Organism(s) | Ref.§ |
|---|---|---|---|
| Oxytocin |
CYIQNCPLG* CYIQNCPPG* |
Eutherian mammals Monkeys |
P01178 G0ZNF6 |
| Valitocin | CYIQNCPVG* | Squalus acanthias | P43000 |
| Aspartocin | CYINNCPLG* | Squalus acanthias | P42999 |
| Asvatocin | CYINNCPVG* | Cartilaginous fish | P42996 |
| Phasitocin | CYFNNCPIG* | Triakis scyllium | Q6F4F6 |
| Phasvatocin | CYFNNCPVG* | Scyliorhinus caniculus | P42997 |
| Cephalotocin | CYFRNCPIG* | Octopus vulgaris | P80027 |
| Octopressin | CFWTSCPIG* | Octopus vulgaris | Q76DN1 |
| Seritocin | CYIQSCPIG* | Amietophrynus regularis | P42995 |
| Glumitocin | CYISNCPQG* | Cartilaginous fish | P42994 |
| Mesotocin |
CYIQNCPIG* CYFQNCPIG* |
Lungfishes and marsupials Setonix brachyurus |
P08162 0909223A¶ |
| Val4-mesotocin | CYIVNCPIG* | Plethodon shermani | A5A4K4 |
| Phe-mesotocin | CFIQNCPIG* | Neoceratodus forsteri | O57388 |
| Isotocin | CYISNCPIG* | Osteichthyes | P42993 |
| Annetocin# | CFVRNCPTG* | Eisenia foetida | P42998 |
| Vasotocin (ancestral)†† | CYIQNCPRG* | Nonmammalian vertebrates | P69129 |
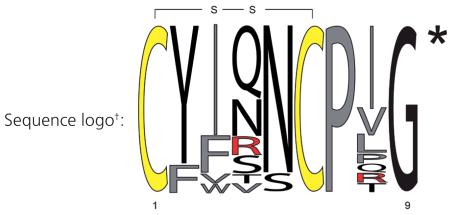
Relative frequency sequence logo plot, conserved cysteine residues are highlighted in yellow, hydrophobic, aromatic and non-polar amino acids are labeled in gray and positively charged residues are labeled in red. All sequences are amidated at the C-terminus, as indicated by the asterisk.
Conserved cysteine residues forming a disulfide bond are highlighted in bold. All sequences are amidated at the C-terminus, as indicated by the asterisk.
UniProtKB entry, unless otherwise stated.
GenBank protein accession number.
Denoted also as Annepressin [1].
Presumed ancestral oxytocin/vasopressin nonapeptide [2].
